当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
High Coverage Profiling of Carboxylated Metabolites in HepG2 Cells Using Secondary Amine-Assisted Ultrahigh-Performance Liquid Chromatography Coupled to High-Resolution Mass Spectrometry
Analytical Chemistry ( IF 7.4 ) Pub Date : 2020-12-27 , DOI: 10.1021/acs.analchem.0c04048 Jie Zheng 1 , Ge-Ge Gong 1 , Shu-Jian Zheng 1 , Yu Zhang 1 , Yu-Qi Feng 1, 2
Analytical Chemistry ( IF 7.4 ) Pub Date : 2020-12-27 , DOI: 10.1021/acs.analchem.0c04048 Jie Zheng 1 , Ge-Ge Gong 1 , Shu-Jian Zheng 1 , Yu Zhang 1 , Yu-Qi Feng 1, 2
Affiliation
|
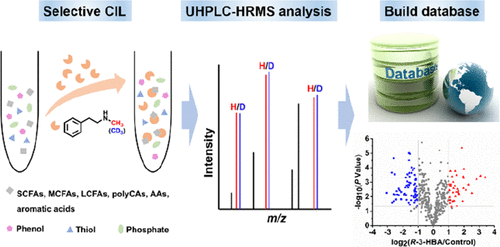
Carboxylic metabolites are an important class of metabolites, which widely exist in mammals with various types. Chemical isotope labeling liquid chromatography-mass spectrometry (CIL-LC-MS) has been widely used for the detection of carboxylated metabolites. However, high coverage analysis of carboxylated metabolites in biological samples is still challenging due to improper reactivity and selectivity of labeling reagents to carboxylated metabolites. In this study, we used N-methylphenylethylamine (MPEA) to label various types of carboxylated metabolites including short-chain fatty acids (SCFAs), medium-chain fatty acids (MCFAs), long-chain fatty acids (LCFAs), polycarboxylic acids (polyCAs), amino acids (AAs), and aromatic acids. Additionally, metabolites containing other functional groups, such as phenol, sulfhydryl, and phosphate groups, could not be labeled under the conditions of MPEA labeling. After MPEA labeling, the detection sensitivity of carboxylic acids was increased by 1–2 orders of magnitude, and their chromatographic retention on a reversed-phase (RP) column was enhanced (RT > 3 min). Under optimized labeling conditions, we used MPEA and d3-N-methylphenylethylamine (d3-MPEA) for high coverage screening of carboxylated metabolites in HepG2 cells by ultrahigh-performance liquid chromatography coupled to high-resolution mass spectrometry (UHPLC-HRMS). As a result, a total of 403 potential carboxylated metabolites were obtained of which 68 were confirmed based on our established in-house chemically labeled metabolite database (CLMD). SCFAs, MCFAs, LCFAs, polyCAs, AAs, and aromatic acids were all detected in HepG2 cell extracts. Due to the successful identification of AAs, the current method increased the coverage of carboxylated metabolites compared with our previous work. Moreover, 133 and 109 carboxylated metabolites with changed contents were obtained in HepG2 cells incubated with curcumin and R-3-hydroxybutyric acid, respectively. In general, our established method realized high coverage analysis of carboxylated metabolites in HepG2 cells.
中文翻译:

二次胺辅助超高效液相色谱与高分辨率质谱联用对HepG2细胞中羧化代谢产物的高覆盖谱分析
羧酸代谢物是一类重要的代谢物,广泛存在于各种类型的哺乳动物中。化学同位素标记液相色谱-质谱(CIL-LC-MS)已被广泛用于检测羧化代谢物。然而,由于标记试剂对羧化代谢物的反应性和选择性不当,生物样品中羧化代谢物的高覆盖率分析仍然具有挑战性。在这项研究中,我们使用N-甲基苯基乙胺(MPEA)标记各种类型的羧化代谢产物,包括短链脂肪酸(SCFA),中链脂肪酸(MCFA),长链脂肪酸(LCFA),多元羧酸(polyCA),氨基酸(AA) )和芳香酸。此外,在MPEA标记的条件下,不能标记含有其他官能团(例如苯酚,巯基和磷酸基)的代谢物。MPEA标记后,羧酸的检测灵敏度提高了1-2个数量级,并且它们在反相(RP)色谱柱上的色谱保留率得到了提高(RT> 3 min)。在优化的标记条件下,我们使用了MPEA和d 3 - N-甲基苯基乙胺(d 3-MPEA),用于通过超高效液相色谱和高分辨率质谱(UHPLC-HRMS)进行HepG2细胞中羧化代谢产物的高覆盖率筛选。结果,总共获得了403种潜在的羧化代谢产物,其中基于我们建立的内部化学标记代谢产物数据库(CLMD)确认了68种。在HepG2细胞提取物中检测到了SCFA,MCFA,LCFA,polyCA,AA和芳香酸。由于成功鉴定了AA,与我们以前的工作相比,当前方法增加了羧化代谢产物的覆盖范围。此外,在用姜黄素和R孵育的HepG2细胞中获得了133和109个含量变化的羧化代谢产物分别为-3-羟基丁酸。通常,我们建立的方法实现了HepG2细胞中羧化代谢产物的高覆盖率分析。
更新日期:2021-01-26
中文翻译:

二次胺辅助超高效液相色谱与高分辨率质谱联用对HepG2细胞中羧化代谢产物的高覆盖谱分析
羧酸代谢物是一类重要的代谢物,广泛存在于各种类型的哺乳动物中。化学同位素标记液相色谱-质谱(CIL-LC-MS)已被广泛用于检测羧化代谢物。然而,由于标记试剂对羧化代谢物的反应性和选择性不当,生物样品中羧化代谢物的高覆盖率分析仍然具有挑战性。在这项研究中,我们使用N-甲基苯基乙胺(MPEA)标记各种类型的羧化代谢产物,包括短链脂肪酸(SCFA),中链脂肪酸(MCFA),长链脂肪酸(LCFA),多元羧酸(polyCA),氨基酸(AA) )和芳香酸。此外,在MPEA标记的条件下,不能标记含有其他官能团(例如苯酚,巯基和磷酸基)的代谢物。MPEA标记后,羧酸的检测灵敏度提高了1-2个数量级,并且它们在反相(RP)色谱柱上的色谱保留率得到了提高(RT> 3 min)。在优化的标记条件下,我们使用了MPEA和d 3 - N-甲基苯基乙胺(d 3-MPEA),用于通过超高效液相色谱和高分辨率质谱(UHPLC-HRMS)进行HepG2细胞中羧化代谢产物的高覆盖率筛选。结果,总共获得了403种潜在的羧化代谢产物,其中基于我们建立的内部化学标记代谢产物数据库(CLMD)确认了68种。在HepG2细胞提取物中检测到了SCFA,MCFA,LCFA,polyCA,AA和芳香酸。由于成功鉴定了AA,与我们以前的工作相比,当前方法增加了羧化代谢产物的覆盖范围。此外,在用姜黄素和R孵育的HepG2细胞中获得了133和109个含量变化的羧化代谢产物分别为-3-羟基丁酸。通常,我们建立的方法实现了HepG2细胞中羧化代谢产物的高覆盖率分析。

























 京公网安备 11010802027423号
京公网安备 11010802027423号