当前位置:
X-MOL 学术
›
Microbiologyopen
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Hidden heterogeneity and co‐occurrence networks of soil prokaryotic communities revealed at the scale of individual soil aggregates
MicrobiologyOpen ( IF 3.4 ) Pub Date : 2020-12-25 , DOI: 10.1002/mbo3.1144 Márton Szoboszlay 1 , Christoph C Tebbe 1
MicrobiologyOpen ( IF 3.4 ) Pub Date : 2020-12-25 , DOI: 10.1002/mbo3.1144 Márton Szoboszlay 1 , Christoph C Tebbe 1
Affiliation
|
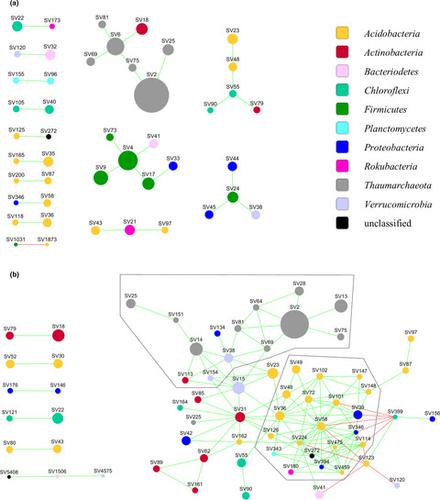
Sequencing PCR‐amplified gene fragments from metagenomic DNA is a widely applied method for studying the diversity and dynamics of soil microbial communities. Typically, DNA is extracted from 0.25 to 1 g of soil. These amounts, however, neglect the heterogeneity of soil present at the scale of soil aggregates and thus ignore a crucial scale for understanding the structure and functionality of soil microbial communities. Here, we show with a nitrogen‐depleted agricultural soil the impact of reducing the amount of soil used for DNA extraction from 250 mg to approx. 1 mg to access spatial information on the prokaryotic community structure, as indicated by 16S rRNA gene amplicon analyses. Furthermore, we demonstrate that individual aggregates from the same soil differ in their prokaryotic community compositions. The analysis of 16S rRNA gene amplicon sequences from individual soil aggregates allowed us, in contrast to 250 mg soil samples, to construct a co‐occurrence network that provides insight into the structure of microbial associations in the studied soil. Two dense clusters were apparent in the network, one dominated by Thaumarchaeota, known to be capable of ammonium oxidation at low N concentrations, and the other by Acidobacteria subgroup 6, representing an oligotrophic lifestyle to obtain energy from SOC. Overall this study demonstrates that DNA obtained from individual soil aggregates provides new insights into how microbial communities are assembled.
中文翻译:

土壤原核生物群落的隐藏异质性和共现网络在单个土壤团聚体的尺度上揭示
从宏基因组 DNA 中对 PCR 扩增的基因片段进行测序是研究土壤微生物群落多样性和动态的一种广泛应用的方法。通常,从 0.25 到 1 克土壤中提取 DNA。然而,这些数量忽略了土壤团聚体尺度上土壤的异质性,因此忽略了理解土壤微生物群落结构和功能的关键尺度。在这里,我们用贫氮的农业土壤展示了将用于 DNA 提取的土壤量从 250 mg 减少到大约 250 mg 的影响。1 mg 以获取有关原核生物群落结构的空间信息,如 16S rRNA 基因扩增子分析所示。此外,我们证明了来自同一土壤的个体聚集体的原核群落组成不同。与 250 mg 土壤样品相比,对来自单个土壤聚集体的 16S rRNA 基因扩增子序列的分析使我们能够构建一个共现网络,从而深入了解所研究土壤中微生物关联的结构。网络中出现了两个密集的集群,一个由Thaumarchaeota,已知能够在低 N 浓度下进行铵氧化,另一个是Acidobacteria亚组 6,代表一种从 SOC 获取能量的贫营养生活方式。总体而言,这项研究表明,从单个土壤聚集体中获得的 DNA 为微生物群落的组装方式提供了新的见解。
更新日期:2021-02-16
中文翻译:

土壤原核生物群落的隐藏异质性和共现网络在单个土壤团聚体的尺度上揭示
从宏基因组 DNA 中对 PCR 扩增的基因片段进行测序是研究土壤微生物群落多样性和动态的一种广泛应用的方法。通常,从 0.25 到 1 克土壤中提取 DNA。然而,这些数量忽略了土壤团聚体尺度上土壤的异质性,因此忽略了理解土壤微生物群落结构和功能的关键尺度。在这里,我们用贫氮的农业土壤展示了将用于 DNA 提取的土壤量从 250 mg 减少到大约 250 mg 的影响。1 mg 以获取有关原核生物群落结构的空间信息,如 16S rRNA 基因扩增子分析所示。此外,我们证明了来自同一土壤的个体聚集体的原核群落组成不同。与 250 mg 土壤样品相比,对来自单个土壤聚集体的 16S rRNA 基因扩增子序列的分析使我们能够构建一个共现网络,从而深入了解所研究土壤中微生物关联的结构。网络中出现了两个密集的集群,一个由Thaumarchaeota,已知能够在低 N 浓度下进行铵氧化,另一个是Acidobacteria亚组 6,代表一种从 SOC 获取能量的贫营养生活方式。总体而言,这项研究表明,从单个土壤聚集体中获得的 DNA 为微生物群落的组装方式提供了新的见解。


























 京公网安备 11010802027423号
京公网安备 11010802027423号