Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2020-12-14 , DOI: 10.1016/j.jbi.2020.103661 Mostafa Akhavan-Safar 1 , Babak Teimourpour 1 , Mehrdad Kargari 1
|
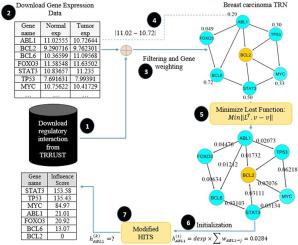
Cancer is among the diseases causing death, in which, cells uncontrollably grow and reproduce beyond the cell regulatory mechanism. In this disease, some genes are initiators of abnormalities and then transmit them to other genes through protein interactions. Accordingly, these genes are known as cancer driver genes (CDGs). In this regard, several methods have been previously developed for identifying cancer driver genes. Most of these methods are computational-based, which use the concept of mutation to predict CDGs. In this research, a method has been proposed for identifying CDGs in the transcription regulatory network using the concept of influence diffusion and by modifying the Hyperlink-Induced Topic Search algorithm based on the diffusion concept. Due to the type of these networks and the processes of abnormality progression in cells and the formation of cancerous tumors, high-influence genes can be the most likely considered as the driver genes. Therefore, we can use the influence diffusion concept as an acceptable theory to identify these genes. Recently, a method has been proposed to detect CDGs with the concept of the influence maximization. One of the challenges in these types of networks is finding the power of regulatory interaction between genes. Moreover, we have proposed a novel method to calculate the weight of regulatory interactions, based on the concept of diffusion. The performance of the proposed method was compared with other seventeen computational and network tools. Correspondingly, three cancer types were used as benchmarks as follows: breast invasive carcinoma (BRCA), Colon adenocarcinoma (COAD), and lung squamous cell carcinoma (LUSC). In addition, to determine the accuracy of the detected drivers using each method, CGC (Cancer Gene Census) and Mut-driver gene lists were utilized as gold standard. The results show that GenHITS performs better compared to the most of the other computational and network methods. Besides, it is also able to identify genes that have been identified by none of the other methods yet.
中文翻译:

GenHITS:一种通过基因影响力评估在人类调节网络中进行驱动基因检测的网络科学方法
癌症是导致死亡的疾病之一,在这种疾病中,细胞无法控制地生长和繁殖,超出了细胞调节机制。在这种疾病中,某些基因是异常的始作俑者,然后通过蛋白质相互作用将它们传递给其他基因。因此,这些基因被称为癌症驱动基因(CDG)。在这方面,先前已经开发了几种用于鉴定癌症驱动基因的方法。这些方法大多数基于计算,使用突变的概念来预测CDG。在这项研究中,已经提出了一种使用影响扩散的概念并通过修改基于扩散概念的超链接诱导主题搜索算法来识别转录调控网络中CDG的方法。由于这些网络的类型以及细胞中异常进展的过程以及癌性肿瘤的形成,高影响力基因最有可能被视为驱动基因。因此,我们可以使用影响扩散的概念作为可接受的理论来鉴定这些基因。近来,已经提出了一种以影响最大化的概念来检测CDG的方法。这些类型网络中的挑战之一是发现基因之间调节相互作用的力量。此外,基于扩散的概念,我们提出了一种新的方法来计算调节相互作用的权重。将该方法的性能与其他十七种计算和网络工具进行了比较。相应地,以下三种癌症类型被用作基准:乳腺癌,乳腺癌,结肠腺癌(COAD)和肺鳞状细胞癌(LUSC)。另外,为了确定使用每种方法检测到的驱动程序的准确性,将CGC(癌症基因普查)和Mut-driver基因列表用作黄金标准。结果表明,与大多数其他计算和网络方法相比,GenHITS的性能更好。此外,它还能够鉴定尚未通过其他方法鉴定的基因。


























 京公网安备 11010802027423号
京公网安备 11010802027423号