当前位置:
X-MOL 学术
›
Microbiologyopen
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Epipelagic microbiome of the Small Aral Sea: Metagenomic structure and ecological diversity
MicrobiologyOpen ( IF 3.4 ) Pub Date : 2020-12-11 , DOI: 10.1002/mbo3.1142 Madina Alexyuk 1 , Andrey Bogoyavlenskiy 1 , Pavel Alexyuk 1 , Yergali Moldakhanov 1 , Vladimir Berezin 1 , Ilya Digel 2
MicrobiologyOpen ( IF 3.4 ) Pub Date : 2020-12-11 , DOI: 10.1002/mbo3.1142 Madina Alexyuk 1 , Andrey Bogoyavlenskiy 1 , Pavel Alexyuk 1 , Yergali Moldakhanov 1 , Vladimir Berezin 1 , Ilya Digel 2
Affiliation
|
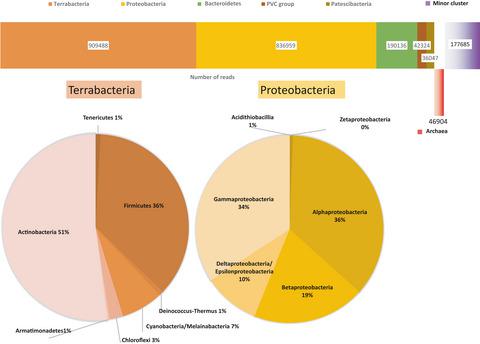
Microbial diversity studies regarding the aquatic communities that experienced or are experiencing environmental problems are essential for the comprehension of the remediation dynamics. In this pilot study, we present data on the phylogenetic and ecological structure of microorganisms from epipelagic water samples collected in the Small Aral Sea (SAS). The raw data were generated by massive parallel sequencing using the shotgun approach. As expected, most of the identified DNA sequences belonged to Terrabacteria and Actinobacteria (40% and 37% of the total reads, respectively). The occurrence of Deinococcus‐Thermus, Armatimonadetes, Chloroflexi in the epipelagic SAS waters was less anticipated. Surprising was also the detection of sequences, which are characteristic for strict anaerobes—Ignavibacteria, hydrogen‐oxidizing bacteria, and archaeal methanogenic species. We suppose that the observed very broad range of phylogenetic and ecological features displayed by the SAS reads demonstrates a more intensive mixing of water masses originating from diverse ecological niches of the Aral‐Syr Darya River basin than presumed before.
中文翻译:

小咸海的表层微生物组:宏基因组结构和生态多样性
关于经历过或正在经历环境问题的水生群落的微生物多样性研究对于理解修复动态至关重要。在这项试点研究中,我们提供了小咸海 (SAS) 上层水样中微生物的系统发育和生态结构数据。原始数据是使用鸟枪法通过大规模并行测序生成的。正如预期的那样,大多数已识别的 DNA 序列属于Terrabacteria和Actinobacteria(分别占总读数的 40% 和 37%)。在上层 SAS 水域中,奇球菌-栖热菌、Artimonadetes和Chloroflexi的出现是不太预料到的。令人惊讶的是序列的检测,这是严格厌氧菌(Ignavibacteria、氢氧化细菌和古菌产甲烷物种)的特征。我们认为,SAS 读数所显示的观察到的非常广泛的系统发育和生态特征表明,来自咸锡尔河流域不同生态位的水团的混合比之前的假设更加密集。
更新日期:2021-02-15
中文翻译:

小咸海的表层微生物组:宏基因组结构和生态多样性
关于经历过或正在经历环境问题的水生群落的微生物多样性研究对于理解修复动态至关重要。在这项试点研究中,我们提供了小咸海 (SAS) 上层水样中微生物的系统发育和生态结构数据。原始数据是使用鸟枪法通过大规模并行测序生成的。正如预期的那样,大多数已识别的 DNA 序列属于Terrabacteria和Actinobacteria(分别占总读数的 40% 和 37%)。在上层 SAS 水域中,奇球菌-栖热菌、Artimonadetes和Chloroflexi的出现是不太预料到的。令人惊讶的是序列的检测,这是严格厌氧菌(Ignavibacteria、氢氧化细菌和古菌产甲烷物种)的特征。我们认为,SAS 读数所显示的观察到的非常广泛的系统发育和生态特征表明,来自咸锡尔河流域不同生态位的水团的混合比之前的假设更加密集。


























 京公网安备 11010802027423号
京公网安备 11010802027423号