Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2020-12-03 , DOI: 10.1016/j.jbi.2020.103652 Maxwell Salvatore 1 , Lauren J Beesley 1 , Lars G Fritsche 2 , David Hanauer 3 , Xu Shi 1 , Alison M Mondul 4 , Celeste Leigh Pearce 4 , Bhramar Mukherjee 1
|
Background
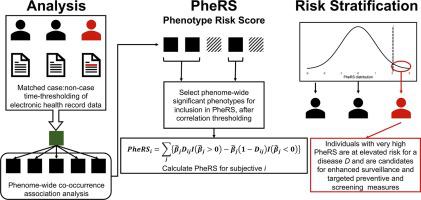
Traditional methods for disease risk prediction and assessment, such as diagnostic tests using serum, urine, blood, saliva or imaging biomarkers, have been important for identifying high-risk individuals for many diseases, leading to early detection and improved survival. For pancreatic cancer, traditional methods for screening have been largely unsuccessful in identifying high-risk individuals in advance of disease progression leading to high mortality and poor survival. Electronic health records (EHR) linked to genetic profiles provide an opportunity to integrate multiple sources of patient information for risk prediction and stratification. We leverage a constellation of temporally associated diagnoses available in the EHR to construct a summary risk score, called a phenotype risk score (PheRS), for identifying individuals at high-risk for having pancreatic cancer. The proposed PheRS approach incorporates the time with respect to disease onset into the prediction framework. We combine and contrast the PheRS with more well-known measures of inherited susceptibility, namely, the polygenic risk scores (PRS) for prediction of pancreatic cancer.
Methodology
We first calculated pairwise, unadjusted associations between pancreatic cancer diagnosis and all possible other diagnoses across the medical phenome. We call these pairwise associations co-occurrences. After accounting for cross-phenotype correlations, the multivariable association estimates from a subset of relatively independent diagnoses were used to create a weighted sum PheRS. We constructed time-restricted risk scores using data from 38,359 participants in the Michigan Genomics Initiative (MGI) based on the diagnoses contained in the EHR at 0, 1, 2, and 5 years prior to the target pancreatic cancer diagnosis. The PheRS was assessed for predictability in the UK Biobank (UKB). We tested the relative contribution of PheRS when added to a model containing a summary measure of inherited genetic susceptibility (PRS) plus other covariates like age, sex, smoking status, drinking status, and body mass index (BMI).
Results
Our exploration of co-occurrence patterns identified expected associations while also revealing unexpected relationships that may warrant closer attention. Solely using the pancreatic cancer PheRS at 5 years before the target diagnoses yielded an AUC of 0.60 (95% CI = [0.58, 0.62]) in UKB. A larger predictive model including PheRS, PRS, and the covariates at the 5-year threshold achieved an AUC of 0.74 (95% CI = [0.72, 0.76]) in UKB. We note that PheRS does contribute independently in the joint model. Finally, scores at the top percentiles of the PheRS distribution demonstrated promise in terms of risk stratification. Scores in the top 2% were 10.20 (95% CI = [9.34, 12.99]) times more likely to identify cases than those in the bottom 98% in UKB at the 5-year threshold prior to pancreatic cancer diagnosis.
Conclusions
We developed a framework for creating a time-restricted PheRS from EHR data for pancreatic cancer using the rich information content of a medical phenome. In addition to identifying hypothesis-generating associations for future research, this PheRS demonstrates a potentially important contribution in identifying high-risk individuals, even after adjusting for PRS for pancreatic cancer and other traditional epidemiologic covariates. The methods are generalizable to other phenotypic traits.
中文翻译:

使用带时间戳的电子健康记录数据进行胰腺癌表型风险评分 (PheRS):在两个大型生物库中的发现和验证
背景
疾病风险预测和评估的传统方法,例如使用血清、尿液、血液、唾液或成像生物标志物的诊断测试,对于识别许多疾病的高风险个体非常重要,从而实现早期发现和提高生存率。对于胰腺癌,传统的筛查方法在疾病进展之前识别高风险个体方面基本上不成功,导致高死亡率和低生存率。与基因图谱相关的电子健康记录 (EHR) 提供了整合多个患者信息来源以进行风险预测和分层的机会。我们利用 EHR 中提供的一系列时间相关诊断来构建一个汇总风险评分,称为表型风险评分 (PheRS),用于识别患有胰腺癌的高风险个体。所提出的 PheRS 方法将疾病发作的时间纳入预测框架。我们将 PheRS 与更知名的遗传易感性指标(即用于预测胰腺癌的多基因风险评分 (PRS))相结合并进行对比。
方法
我们首先计算了胰腺癌诊断与医学现象中所有可能的其他诊断之间的成对、未经调整的关联。我们将这些成对关联称为共现。在考虑了跨表型相关性后,使用来自相对独立的诊断子集的多变量关联估计来创建加权和 PheRS。我们根据 EHR 中包含的目标胰腺癌诊断前0、1、2 和 5 年的诊断,使用密歇根基因组计划 (MGI) 38,359 名参与者的数据构建了时间限制的风险评分。英国生物银行 (UKB) 对 PheRS 的可预测性进行了评估。我们测试了将 PheRS 添加到包含遗传易感性 (PRS) 汇总测量以及年龄、性别、吸烟状况、饮酒状况和体重指数 (BMI) 等其他协变量的模型中时的相对贡献。
结果
我们对共现模式的探索确定了预期的关联,同时也揭示了可能值得密切关注的意外关系。在目标诊断前 5 年单独使用胰腺癌 PheRS,UKB 中的 AUC 为 0.60(95% CI = [0.58, 0.62])。包括 PheRS、PRS 和 5 年阈值协变量在内的更大预测模型在 UKB 中的 AUC 为 0.74(95% CI = [0.72, 0.76])。我们注意到 PheRS 在联合模型中确实做出了独立贡献。最后,PheRS 分布最高百分位数的分数证明了风险分层的前景。在诊断胰腺癌之前的 5 年阈值中,UKB 中得分最高的 2% 的人识别病例的可能性是得分最低的 98% 的人的 10.20 (95% CI = [9.34, 12.99]) 倍。
结论
我们开发了一个框架,利用医学现象的丰富信息内容,根据胰腺癌的 EHR 数据创建有时间限制的 PheRS。除了为未来研究确定假设生成关联之外,即使在调整了胰腺癌和其他传统流行病学协变量的 PRS 后,该 PheRS 还证明了在识别高风险个体方面的潜在重要贡献。这些方法可推广到其他表型性状。



























 京公网安备 11010802027423号
京公网安备 11010802027423号