当前位置:
X-MOL 学术
›
NMR Biomed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Voxel‐wise partial volume correction method for accurate estimation of tissue sodium concentration in 23Na‐MRI at 7 T
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2020-12-03 , DOI: 10.1002/nbm.4448 Sang-Young Kim 1 , Junghyun Song 1 , Jong-Hyun Yoon 1 , Kyoung-Nam Kim 2 , Jun-Young Chung 1, 3 , Young Noh 1, 4
NMR in Biomedicine ( IF 2.9 ) Pub Date : 2020-12-03 , DOI: 10.1002/nbm.4448 Sang-Young Kim 1 , Junghyun Song 1 , Jong-Hyun Yoon 1 , Kyoung-Nam Kim 2 , Jun-Young Chung 1, 3 , Young Noh 1, 4
Affiliation
|
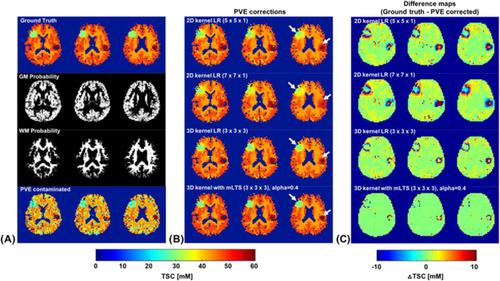
Sodium is crucial for the maintenance of cell physiology, and its regulation of the sodium‐potassium pump has implications for various neurological conditions. The distribution of sodium concentrations in tissue can be quantitatively evaluated by means of sodium MRI (23Na‐MRI). Despite its usefulness in diagnosing particular disease conditions, tissue sodium concentration (TSC) estimated from 23Na‐MRI can be strongly biased by partial volume effects (PVEs) that are induced by broad point spread functions (PSFs) as well as tissue fraction effects. In this work, we aimed to propose a robust voxel‐wise partial volume correction (PVC) method for 23Na‐MRI. The method is based on a linear regression (LR) approach to correct for tissue fraction effects, but it utilizes a 3D kernel combined with a modified least trimmed square (3D‐mLTS) method in order to minimize regression‐induced inherent smoothing effects. We acquired 23Na‐MRI data with conventional Cartesian sampling at 7 T, and spill‐over effects due to the PSF were considered prior to correcting for tissue fraction effects using 3D‐mLTS. In the simulation, we found that the TSCs of gray matter (GM) and white matter (WM) were underestimated by 20% and 11% respectively without correcting tissue fraction effects, but the differences between ground truth and PVE‐corrected data after the PVC using the 3D‐mLTS method were only approximately 0.6% and 0.4% for GM and WM, respectively. The capability of the 3D‐mLTS method was further demonstrated with in vivo 23Na‐MRI data, showing significantly lower regression errors (ie root mean squared error) as compared with conventional LR methods (p < 0.001). The results of simulation and in vivo experiments revealed that 3D‐mLTS is superior for determining under‐ or overestimated TSCs while preserving anatomical details. This suggests that the 3D‐mLTS method is well suited for the accurate determination of TSC, especially in small focal lesions associated with pathological conditions.
中文翻译:

体素部分体积校正方法用于准确估计 23Na-MRI 在 7 T 下的组织钠浓度
钠对于维持细胞生理机能至关重要,它对钠钾泵的调节对各种神经系统疾病都有影响。可以通过钠 MRI ( 23 Na-MRI) 定量评估组织中钠浓度的分布。尽管它在诊断特定疾病状况方面有用,但从23 Na-MRI 估计的组织钠浓度 (TSC) 可能会受到由宽点扩散函数 (PSF) 和组织分数效应引起的部分体积效应 (PVE) 的强烈偏差。在这项工作中,我们旨在为23钠磁共振成像。该方法基于线性回归 (LR) 方法来校正组织分数效应,但它利用 3D 内核与改进的最小修剪平方 (3D-mLTS) 方法相结合,以最大限度地减少回归引起的固有平滑效应。我们获得了23在使用 3D-mLTS 校正组织分数效应之前,考虑了在 7 T 下使用传统笛卡尔采样的 Na-MRI 数据,以及由 PSF 引起的溢出效应。在模拟中,我们发现在没有校正组织分数效应的情况下,灰质 (GM) 和白质 (WM) 的 TSC 分别被低估了 20% 和 11%,但在 PVC 之后,ground truth 和 PVE 校正数据之间存在差异使用 3D-mLTS 方法的 GM 和 WM 分别只有大约 0.6% 和 0.4%。体内23 Na-MRI 数据进一步证明了 3D-mLTS 方法的能力,与传统的 LR 方法相比,回归误差(即均方根误差)显着降低(p< 0.001)。模拟和体内实验的结果表明,3D-mLTS 在确定低估或高估的 TSCs 方面具有优越性,同时保留了解剖细节。这表明 3D-mLTS 方法非常适合准确测定 TSC,特别是在与病理状况相关的小局灶性病变中。
更新日期:2021-01-04
中文翻译:

体素部分体积校正方法用于准确估计 23Na-MRI 在 7 T 下的组织钠浓度
钠对于维持细胞生理机能至关重要,它对钠钾泵的调节对各种神经系统疾病都有影响。可以通过钠 MRI ( 23 Na-MRI) 定量评估组织中钠浓度的分布。尽管它在诊断特定疾病状况方面有用,但从23 Na-MRI 估计的组织钠浓度 (TSC) 可能会受到由宽点扩散函数 (PSF) 和组织分数效应引起的部分体积效应 (PVE) 的强烈偏差。在这项工作中,我们旨在为23钠磁共振成像。该方法基于线性回归 (LR) 方法来校正组织分数效应,但它利用 3D 内核与改进的最小修剪平方 (3D-mLTS) 方法相结合,以最大限度地减少回归引起的固有平滑效应。我们获得了23在使用 3D-mLTS 校正组织分数效应之前,考虑了在 7 T 下使用传统笛卡尔采样的 Na-MRI 数据,以及由 PSF 引起的溢出效应。在模拟中,我们发现在没有校正组织分数效应的情况下,灰质 (GM) 和白质 (WM) 的 TSC 分别被低估了 20% 和 11%,但在 PVC 之后,ground truth 和 PVE 校正数据之间存在差异使用 3D-mLTS 方法的 GM 和 WM 分别只有大约 0.6% 和 0.4%。体内23 Na-MRI 数据进一步证明了 3D-mLTS 方法的能力,与传统的 LR 方法相比,回归误差(即均方根误差)显着降低(p< 0.001)。模拟和体内实验的结果表明,3D-mLTS 在确定低估或高估的 TSCs 方面具有优越性,同时保留了解剖细节。这表明 3D-mLTS 方法非常适合准确测定 TSC,特别是在与病理状况相关的小局灶性病变中。


























 京公网安备 11010802027423号
京公网安备 11010802027423号