当前位置:
X-MOL 学术
›
J. Phys. Chem. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Degree of N-Methylation of Nucleosides and Metabolites Controls Binding Affinity to Pristine Silica Surfaces
The Journal of Physical Chemistry Letters ( IF 5.7 ) Pub Date : 2020-11-30 , DOI: 10.1021/acs.jpclett.0c02888 Mouzhe Xie 1 , Rafael Brüschweiler 1, 2, 3
The Journal of Physical Chemistry Letters ( IF 5.7 ) Pub Date : 2020-11-30 , DOI: 10.1021/acs.jpclett.0c02888 Mouzhe Xie 1 , Rafael Brüschweiler 1, 2, 3
Affiliation
|
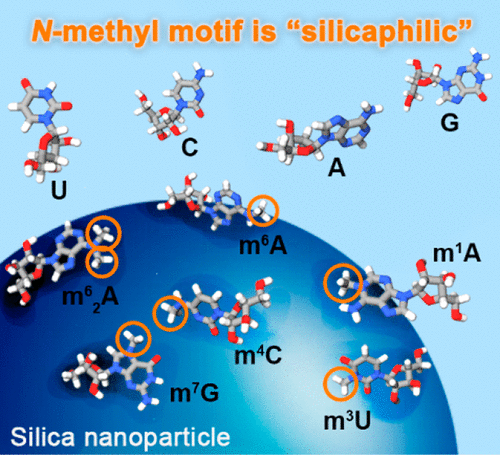
Biological molecules interact with silica (SiO2) surfaces with binding affinities that greatly vary depending on their physical–chemical properties. However, the quantitative characterization of biological compounds adsorbed on silica surfaces, especially of compounds involved in fast, reversible interactions, has been challenging, and the driving forces are not well understood. Here, we show how carbon-13 NMR spin relaxation provides quantitative atomic-detail information about the transient molecular binding to pristine silica surfaces, represented by colloidally dispersed silica nanoparticles (SNPs). Based on the quantitative analysis of almost two dozen biological molecules, we find that the addition of N-methyl motifs systematically increases molecular binding affinities to silica in a nearly quantitatively predictable manner. Among the studied compounds are methylated nucleosides, which are common in epigenetic signaling in nucleic acids. The quantitative understanding of N-methylation may open up new ways to detect and separate methylated nucleic acids or even regulate their cellular functions.
中文翻译:

核苷和代谢物的N-甲基化程度控制与原始二氧化硅表面的结合亲和力
生物分子与二氧化硅(SiO 2)表面相互作用,其结合亲和力根据其理化性质而有很大差异。然而,吸附在二氧化硅表面上的生物化合物,特别是参与快速,可逆相互作用的化合物的定量表征一直具有挑战性,并且对驱动力的了解还不够。在这里,我们展示了碳13 NMR自旋弛豫如何提供有关原始分子硅胶表面瞬态分子结合的定量原子细节信息,以胶态分散的二氧化硅纳米颗粒(SNPs)表示。通过对近两打生物分子的定量分析,我们发现氮的添加-甲基基序以几乎定量可预测的方式系统地增加了与二氧化硅的分子结合亲和力。在研究的化合物中有甲基化的核苷,它们在核酸的表观遗传信号中很常见。对N-甲基化的定量理解可以开辟新的方法来检测和分离甲基化的核酸,甚至调节其细胞功能。
更新日期:2020-12-17
中文翻译:

核苷和代谢物的N-甲基化程度控制与原始二氧化硅表面的结合亲和力
生物分子与二氧化硅(SiO 2)表面相互作用,其结合亲和力根据其理化性质而有很大差异。然而,吸附在二氧化硅表面上的生物化合物,特别是参与快速,可逆相互作用的化合物的定量表征一直具有挑战性,并且对驱动力的了解还不够。在这里,我们展示了碳13 NMR自旋弛豫如何提供有关原始分子硅胶表面瞬态分子结合的定量原子细节信息,以胶态分散的二氧化硅纳米颗粒(SNPs)表示。通过对近两打生物分子的定量分析,我们发现氮的添加-甲基基序以几乎定量可预测的方式系统地增加了与二氧化硅的分子结合亲和力。在研究的化合物中有甲基化的核苷,它们在核酸的表观遗传信号中很常见。对N-甲基化的定量理解可以开辟新的方法来检测和分离甲基化的核酸,甚至调节其细胞功能。


























 京公网安备 11010802027423号
京公网安备 11010802027423号