International Journal of Medical Informatics ( IF 4.9 ) Pub Date : 2020-11-27 , DOI: 10.1016/j.ijmedinf.2020.104348 K. Sepehri , X. Song , R. Proulx , S. Ghosh Hajra , B. Dobberthien , C.C. Liu , R.C.N. D’Arcy , D. Murray , A.V. Krauze
|
Purpose/objective(s)
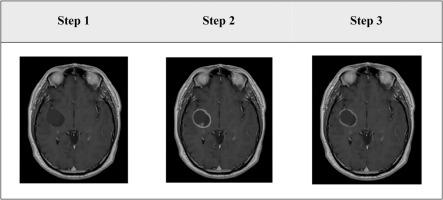
Gliomas are uniformly fatal brain tumours with significant neurological and quality of life detriment to patients. Improvement in outcomes has remained largely unchanged in nearly 20 years. MRI (magnetic resonance imaging) is often used in diagnosis and management. Machine learning analyses of large-scale MRI data are pivotal in advancing the diagnosis, management and improve outcomes in neuro-oncology. A common challenge to robust machine learning approaches is the lack of large ‘ground truth’ datasets in supervised learning for building classification and prediction models. The creation of these datasets relies on human-expert input and is time-consuming and subjective error-prone, limiting effective machine learning applications. Simulation of mechanistic aspects such as geometry, location and physical properties of brain tumours can generate large-scale ground-truth datasets allowing for comparison of analysis techniques in clinical applications. We aimed to develop a transparent and convenient method for building ‘ground truth’ presentations of simulated glioma lesions on anatomical MRI.
Materials/methods
The simulation workflow was created using the Feature Manipulation Engine (FME®), a data integration platform specializing in the spatial data processing. By compiling and integrating FME’s functions to read, integrate, transform, validate, save, and display MRI data, and experimenting with ways to manipulate the parameters concerning location, size, shape, and signal intensity with the presentations of glioma, we were able to generate simulated appearances of high-grade gliomas on gadolinium-based high-resolution 3D T1-weighted MRI (1 mm3). Data of patients with canonical high-grade tumours were used as real-world tumours for validating the accuracy of the simulation. Twenty raters who are experienced with brain tumour interpretation on MRI independently completed a survey, designed to distinguish simulated and real-world brain tumours. Sensitivity and specificity were calculated for assessing the performance of the approach with the binary classification of simulated vs real-world tumours. Correlation and regression were used in run time analysis, assessing the software toolset’s efficiency in producing different numbers of simulated lesions. Differences in the group means were examined using the non-parametric Kruskal-Wallis test.
Results
The simulation method was developed as an interpretable and useful workflow for the easy creation of tumour simulations and incorporation into 3D MRI. A linear increase in the running time and memory usage was observed with an increasing number of generated lesions. The respondents' accuracy rate ranged between 33.3 and 83.3 %. The sensitivity and specificity were low for a human expert to differentiate simulated lesions from real gliomas (0.43 and 0.58) or vice versa (0.65 and 0.62). The mean scores ranking the real-world gliomas did not differ between the simulated and real tumours.
Conclusion
The reliable and user-friendly software method can allow for robust simulation of high-grade glioma on MRI. Ongoing research efforts include optimizing the workflow for generating glioma datasets as well as adapting it to simulating additional MRI brain changes.
中文翻译:

迈向医学成像分析中的有效机器学习:MRI上胶质瘤“地面真相”模拟的新方法和专家评估
目的/目的
神经胶质瘤是致命的均一性致命的脑肿瘤,对患者的神经和生活质量都有重大影响。近20年以来,结果的改善在很大程度上保持不变。MRI(磁共振成像)通常用于诊断和管理。大规模MRI数据的机器学习分析对于推进神经肿瘤学的诊断,管理和改善结果至关重要。鲁棒的机器学习方法的一个常见挑战是,在用于建筑物分类和预测模型的监督学习中缺少大型的“地面真理”数据集。这些数据集的创建依赖于人类专家的输入,既费时又主观容易出错,从而限制了有效的机器学习应用程序。模拟机械方面,例如几何,脑肿瘤的位置和物理特性可以生成大规模的真实数据集,从而可以在临床应用中比较分析技术。我们旨在开发一种透明,方便的方法,以在解剖MRI上构建模拟神经胶质瘤病变的“地面真相”表现。
材料/方法
仿真工作流是使用功能操纵引擎(FME®)创建的,该引擎是专门用于空间数据处理的数据集成平台。通过编译和集成FME的功能以读取,集成,转换,验证,保存和显示MRI数据,并尝试以胶质瘤的表现方式操纵有关位置,大小,形状和信号强度的参数的方法,我们能够在基于g的高分辨率3D T1加权MRI(1 mm 3)。具有规范性高级别肿瘤患者的数据被用作真实世界的肿瘤,以验证模拟的准确性。二十位在MRI上具有脑肿瘤解释经验的评估者独立完成了一项调查,旨在区分模拟和现实世界中的脑肿瘤。计算敏感性和特异性,以评估模拟肿瘤与真实肿瘤的二元分类方法的效果。相关性和回归用于运行时分析,评估软件工具集产生不同数量的模拟病变的效率。使用非参数Kruskal-Wallis检验检查组平均值的差异。
结果
仿真方法被开发为可解释且有用的工作流程,可轻松创建肿瘤仿真并将其纳入3D MRI。随着产生的病变数量的增加,观察到运行时间和内存使用率呈线性增加。受访者的准确率介于33.3%和83.3%之间。对于人类专家而言,区分模拟病变与真实神经胶质瘤(0.43和0.58),反之亦然(0.65和0.62)的敏感性和特异性较低。真实世界神经胶质瘤的平均评分在模拟肿瘤和真实肿瘤之间没有差异。
结论
可靠且用户友好的软件方法可以在MRI上对高级神经胶质瘤进行可靠的仿真。正在进行的研究工作包括优化用于生成神经胶质瘤数据集的工作流程以及使其适应模拟其他MRI脑部变化的工作流程。


























 京公网安备 11010802027423号
京公网安备 11010802027423号