当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Open Search Strategy for Inferring the Masses of Cross-Link Adducts on Proteins
Analytical Chemistry ( IF 7.4 ) Pub Date : 2020-11-25 , DOI: 10.1021/acs.analchem.0c03292 Moriya Slavin 1 , Tamar Tayri-Wilk 1, 2 , Hala Milhem 1 , Nir Kalisman 1
Analytical Chemistry ( IF 7.4 ) Pub Date : 2020-11-25 , DOI: 10.1021/acs.analchem.0c03292 Moriya Slavin 1 , Tamar Tayri-Wilk 1, 2 , Hala Milhem 1 , Nir Kalisman 1
Affiliation
|
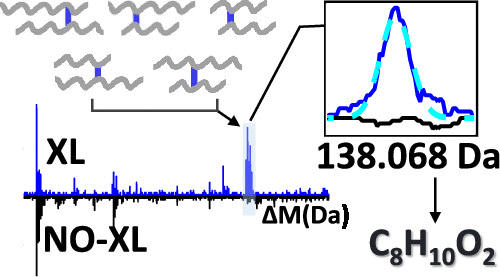
Development of new reagents for protein cross-linking is constantly ongoing. The chemical formulas for the linker adducts formed by these reagents are usually deduced from expert knowledge and then validated by mass spectrometry. Clearly, it would be more rigorous to infer the chemical compositions of the adducts directly from the data without any prior assumptions on their chemistries. Unfortunately, the analysis tools that are currently available to detect chemical modifications on linear peptides are not applicable to the case of two cross-linked peptides. Here, we show that an adaptation of the open search strategy that works on linear peptides can be used to characterize cross-link modifications in pairs of peptides. We benchmark our approach by correctly inferring the linker masses of two well-known reagents, DSS and formaldehyde, to accuracies of a few parts per million. We then investigate the cross-linking chemistries of two poorly characterized reagents: EMCS and glutaraldehyde. In the case of EMCS, we find that the expected cross-linking chemistry is accompanied by a competing chemistry that targets other amino acid types. In the case of glutaraldehyde, we find that the chemical formula of the dominant linker is C5H4, which indicates a ringed aromatic structure. These results demonstrate how, with very little effort, our approach can yield nontrivial insights to better characterize new cross-linkers.
中文翻译:

推断蛋白质交联加合物质量的开放搜索策略
用于蛋白质交联的新试剂的开发一直在进行。由这些试剂形成的接头加合物的化学式通常是根据专业知识推导出来的,然后通过质谱法进行验证。显然,直接从数据中推断加合物的化学组成会更加严格,而无需事先对其化学性质进行任何假设。不幸的是,当前可用于检测线性肽的化学修饰的分析工具不适用于两种交联肽的情况。在这里,我们表明对线性肽起作用的开放搜索策略的改编可用于表征肽对中的交联修饰。我们通过正确推断两种著名试剂DSS和甲醛的连接基团质量来对我们的方法进行基准测试,精确到百万分之几。然后,我们研究了两种表征较差的试剂:EMCS和戊二醛的交联化学。对于EMCS,我们发现预期的交联化学伴随着针对其他氨基酸类型的竞争化学。对于戊二醛,我们发现主要接头的化学式为C5 H 4,其表示环状的芳族结构。这些结果表明,只需很少的努力,我们的方法如何就能获得重要的见解,从而更好地表征新的交联剂。
更新日期:2020-12-15
中文翻译:

推断蛋白质交联加合物质量的开放搜索策略
用于蛋白质交联的新试剂的开发一直在进行。由这些试剂形成的接头加合物的化学式通常是根据专业知识推导出来的,然后通过质谱法进行验证。显然,直接从数据中推断加合物的化学组成会更加严格,而无需事先对其化学性质进行任何假设。不幸的是,当前可用于检测线性肽的化学修饰的分析工具不适用于两种交联肽的情况。在这里,我们表明对线性肽起作用的开放搜索策略的改编可用于表征肽对中的交联修饰。我们通过正确推断两种著名试剂DSS和甲醛的连接基团质量来对我们的方法进行基准测试,精确到百万分之几。然后,我们研究了两种表征较差的试剂:EMCS和戊二醛的交联化学。对于EMCS,我们发现预期的交联化学伴随着针对其他氨基酸类型的竞争化学。对于戊二醛,我们发现主要接头的化学式为C5 H 4,其表示环状的芳族结构。这些结果表明,只需很少的努力,我们的方法如何就能获得重要的见解,从而更好地表征新的交联剂。



























 京公网安备 11010802027423号
京公网安备 11010802027423号