iScience ( IF 5.8 ) Pub Date : 2020-11-23 , DOI: 10.1016/j.isci.2020.101850 Elyse Geoffroy , Isabelle Gregga , Heather E. Wheeler
|
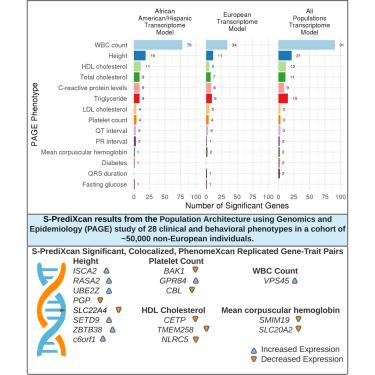
Most genome-wide association studies (GWAS) and transcriptome-wide association studies (TWAS) focus on European populations; however, these results cannot always be accurately applied to non-European populations due to genetic architecture differences. Using GWAS summary statistics in the Population Architecture using Genomics and Epidemiology study, which comprises ∼50,000 Hispanic/Latinos, African Americans, Asians, Native Hawaiians, and Native Americans, we perform TWAS to determine gene-trait associations. We compared results using three transcriptome prediction models derived from Multi-Ethnic Study of Atherosclerosis populations: the African American and Hispanic/Latino (AFHI) model, the European (EUR) model, and the African American, Hispanic/Latino, and European (ALL) model. We identified 240 unique significant trait-associated genes. We found more significant, colocalized genes that replicate in larger cohorts when applying the AFHI model than the EUR or ALL model. Thus, TWAS with population-matched transcriptome models have more power for discovery and replication, demonstrating the need for more transcriptome studies in diverse populations.
中文翻译:

人口匹配的转录组预测增加了TWAS发现和复制率
大多数全基因组和转录组范围的关联研究(GWAS,TWAS)都集中在欧洲人群。但是,由于基因结构的差异,这些结果不能总是准确地应用于非欧洲人口。使用GWAS汇总统计数据进行的使用基因组学和流行病学(PAGE)研究的人口结构研究(包括50,000左右的西班牙裔/拉丁美洲人,非裔美国人,亚洲人,夏威夷原住民和美洲原住民),我们执行TWAS来确定基因-性状关联。我们使用源自多族裔动脉粥样硬化研究(MESA)的三种转录组预测模型比较了结果:非裔美国人和西班牙裔/拉丁美洲人(AFHI)模型,欧洲(EUR)模型以及非裔美国人,西班牙裔/拉丁美洲人和欧洲(ALL)模型。我们确定了240个独特的显着性状相关基因。我们发现,应用AFHI模型时,与EUR或ALL模型相比,在更大的队列中可以复制的更重要的共定位基因。因此,具有群体匹配的转录组模型的TWAS具有更大的发现和复制能力,这表明需要在不同的人群中进行更多的转录组研究。


























 京公网安备 11010802027423号
京公网安备 11010802027423号