Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Accelerating copolymer inverse design using monte carlo tree search
Nanoscale ( IF 6.7 ) Pub Date : 2020-11-14 , DOI: 10.1039/d0nr06091g Tarak K Patra 1 , Troy D Loeffler , Subramanian K R S Sankaranarayanan
Nanoscale ( IF 6.7 ) Pub Date : 2020-11-14 , DOI: 10.1039/d0nr06091g Tarak K Patra 1 , Troy D Loeffler , Subramanian K R S Sankaranarayanan
Affiliation
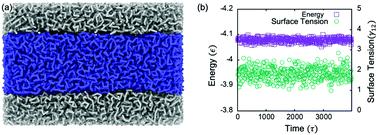
|
There exists a broad class of sequencing problems in soft materials such as proteins and polymers that can be formulated as a heuristic search that involves decision making akin to a computer game. AI gaming algorithms such as Monte Carlo tree search (MCTS) gained prominence after their exemplary performance in the computer Go game and are decision trees aimed at identifying the path (moves) that should be taken by the policy to reach the final winning or optimal solution. Major challenges in inverse sequencing problems are that the materials search space is extremely vast and property evaluation for each sequence is computationally demanding. Reaching an optimal solution by minimizing the total number of evaluations in a given design cycle is therefore highly desirable. We demonstrate that one can adopt this approach for solving the sequencing problem by developing and growing a decision tree, where each node in the tree is a candidate sequence whose fitness is directly evaluated by molecular simulations. We interface MCTS with MD simulations and use a representative example of designing a copolymer compatibilizer, where the goal is to identify sequence specific copolymers that lead to zero interfacial energy between two immiscible homopolymers. We apply the MCTS algorithm to polymer chain lengths varying from 10-mer to 30-mer, wherein the overall search space varies from 210 (1024) to 230 (∼1 billion). In each case, we identify a target sequence that leads to zero interfacial energy within a few hundred evaluations demonstrating the scalability and efficiency of MCTS in exploring practical materials design problems with exceedingly vast chemical/material search space. Our MCTS-MD framework can be easily extended to several other polymer and protein inverse design problems, in particular, for cases where sequence-property data is either unavailable and/or is resource intensive.
中文翻译:

使用蒙特卡罗树搜索加速共聚物逆设计
蛋白质和聚合物等软材料中存在一大类测序问题,这些问题可以表述为涉及类似于计算机游戏的决策的启发式搜索。蒙特卡洛树搜索 (MCTS) 等人工智能游戏算法在计算机围棋游戏中表现出色后获得了广泛关注,它们是决策树,旨在识别策略应采取的路径(走法)以达到最终获胜或最佳解决方案。逆序问题的主要挑战是材料搜索空间极其巨大,并且每个序列的属性评估对计算要求很高。因此,非常需要通过最小化给定设计周期中的评估总数来达到最佳解决方案。我们证明,可以采用这种方法通过开发和生长决策树来解决测序问题,其中树中的每个节点都是候选序列,其适应度通过分子模拟直接评估。我们将 MCTS 与 MD 模拟结合起来,并使用设计共聚物增容剂的代表性示例,其目标是识别序列特定的共聚物,从而使两种不混溶的均聚物之间的界面能为零。我们将 MCTS 算法应用于从 10 聚体到 30 聚体的聚合物链长度,其中整体搜索空间从 2 10 (1024) 到 2 30 (∼10 亿) 变化。在每种情况下,我们都在数百次评估中确定了导致界面能为零的目标序列,证明了 MCTS 在探索具有极其巨大的化学/材料搜索空间的实际材料设计问题时的可扩展性和效率。我们的 MCTS-MD 框架可以轻松扩展到其他几个聚合物和蛋白质逆向设计问题,特别是对于序列特性数据不可用和/或资源密集型的情况。
更新日期:2020-11-21
中文翻译:

使用蒙特卡罗树搜索加速共聚物逆设计
蛋白质和聚合物等软材料中存在一大类测序问题,这些问题可以表述为涉及类似于计算机游戏的决策的启发式搜索。蒙特卡洛树搜索 (MCTS) 等人工智能游戏算法在计算机围棋游戏中表现出色后获得了广泛关注,它们是决策树,旨在识别策略应采取的路径(走法)以达到最终获胜或最佳解决方案。逆序问题的主要挑战是材料搜索空间极其巨大,并且每个序列的属性评估对计算要求很高。因此,非常需要通过最小化给定设计周期中的评估总数来达到最佳解决方案。我们证明,可以采用这种方法通过开发和生长决策树来解决测序问题,其中树中的每个节点都是候选序列,其适应度通过分子模拟直接评估。我们将 MCTS 与 MD 模拟结合起来,并使用设计共聚物增容剂的代表性示例,其目标是识别序列特定的共聚物,从而使两种不混溶的均聚物之间的界面能为零。我们将 MCTS 算法应用于从 10 聚体到 30 聚体的聚合物链长度,其中整体搜索空间从 2 10 (1024) 到 2 30 (∼10 亿) 变化。在每种情况下,我们都在数百次评估中确定了导致界面能为零的目标序列,证明了 MCTS 在探索具有极其巨大的化学/材料搜索空间的实际材料设计问题时的可扩展性和效率。我们的 MCTS-MD 框架可以轻松扩展到其他几个聚合物和蛋白质逆向设计问题,特别是对于序列特性数据不可用和/或资源密集型的情况。


























 京公网安备 11010802027423号
京公网安备 11010802027423号