iScience ( IF 5.8 ) Pub Date : 2020-11-20 , DOI: 10.1016/j.isci.2020.101838 Belinda R. Hauser , Marit H. Aure , Michael C. Kelly , Matthew P. Hoffman , Alejandro M. Chibly
|
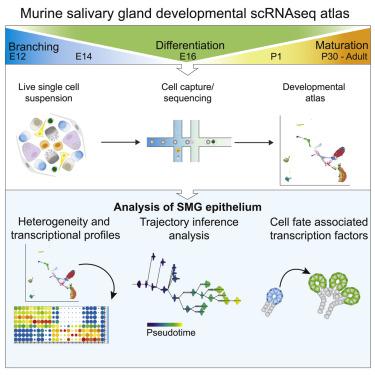
Understanding the dynamic transcriptional landscape throughout organ development will provide a template for regenerative therapies. Here, we generated a single-cell RNA sequencing atlas of murine submandibular glands identifying transcriptional profiles that revealed cellular heterogeneity during landmark developmental events: end bud formation, branching morphogenesis, cytodifferentiation, maturation, and homeostasis. Trajectory inference analysis suggests plasticity among acinar and duct populations. We identify transcription factors correlated with acinar differentiation including Spdef, Etv1, and Xbp1, and loss of Ybx1, Eno1, Sox11, and Atf4. Furthermore, we characterize two intercalated duct populations defined by either Gfra3 and Kit, or Gstt1. This atlas can be used to investigate specific cell functions and comparative studies predicting common mechanisms involved in development of branching organs.
中文翻译:

鼠唾液腺发育单细胞RNAseq地图集的生成。
了解整个器官发育过程中动态转录环境将为再生疗法提供模板。在这里,我们生成了鼠下颌腺的单细胞RNAseq地图集,确定了在具有里程碑意义的发育事件(端芽形成,分支形态发生,细胞分化,成熟和稳态)中揭示细胞异质性的转录谱。轨迹推断分析表明腺泡和导管种群之间具有可塑性。我们确定与腺泡分化相关的转录因子,包括Spdef,Etv1和Xbp1,以及Ybx1,Eno1,Sox11和Atf4的缺失。此外,我们表征了由Gfra3和Kit或Gstt1定义的两个插层导管种群。该图集可用于研究特定的细胞功能,并进行比较研究,预测参与分支器官发育的常见机制。



























 京公网安备 11010802027423号
京公网安备 11010802027423号