International Journal for Parasitology: Parasites and Wildlife ( IF 1.8 ) Pub Date : 2020-11-19 , DOI: 10.1016/j.ijppaw.2020.11.003 S. Keatley , A. Botero , J. Fosu-Nyarko , L. Pallant , A. Northover , R.C.A. Thompson
|
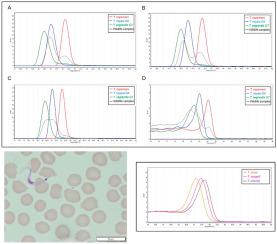
Conventional nested PCR and Sanger sequencing methods are currently the gold standards for detecting trypanosomes in wildlife. However, these techniques are time-consuming and can often overlook mixed infections. True trypanosome prevalence can thus be underrepresented. Here, we designed an 18S rDNA-based real-time quantitative PCR (qPCR) assay coupled with High-Resolution Melting Analysis (HRMA) to detect and discriminate three Trypanosoma species (T. copemani, T. noyesi, and T. vegrandis) commonly infecting Australian marsupials. A total of 68 genetically characterised samples from blood and tissue were used to validate the High-Resolution Melting - Real Time Quantitative Polymerase Chain Reaction (HRM-qPCR) assay. A further 87 marsupial samples consisting of blood, tissue and in vitro cultures derived from wildlife blood samples, were screened for the first time using this assay, and species identity confirmed using conventional PCR and Sanger sequencing. All three Trypanosoma species were successfully detected in pure cultures using the HRM-qPCR assay, and in samples containing mixed trypanosome infections. Of the 87 marsupial samples screened using the HRM-qPCR assay, 93.1% were positive for trypanosomes, and 8.0% contained more than one trypanosome species. In addition to the three targeted Trypanosoma species, this assay was also able to detect and identify other native and exotic trypanosomes. The turnaround time for this assay, from sample preparation to obtaining results, was less than 2 h, with a detection limit of 10 copies of the amplicon in a reaction for each of the targeted trypanosome species. This more rapid and sensitive diagnostic tool provides a high throughput platform for the detection, identification and quantification of trypanosome infections. It will also improve understanding of host diversity and parasite relationships and facilitate conservation management decisions.
中文翻译:

通过高分辨率熔解-实时定量聚合酶链反应(HRM-qPCR)对感染澳大利亚野生动物的锥虫进行物种级鉴定
目前,常规的巢式PCR和Sanger测序方法是检测野生动物中锥虫的金标准。但是,这些技术非常耗时,常常会忽略混合感染。真正的锥虫患病率可能因此被低估。在这里,我们设计了一种基于18S rDNA的实时定量PCR(qPCR)分析方法,并结合了高分辨率熔解分析(HRMA),以检测和区分三种锥虫(T. copemani,T。noyesi和T. vegrandis)通常会感染澳大利亚有袋动物。总共从血液和组织中提取了68个具有遗传特征的样品,用于验证高分辨率熔解-实时定量聚合酶链反应(HRM-qPCR)分析。使用该测定法首次筛选了另外87种有袋动物样品,这些样品由血液,组织和来自野生动物血样的体外培养物组成,并使用常规PCR和Sanger测序确认了物种同一性。三种锥虫使用HRM-qPCR分析法在纯培养物中以及含有混合锥虫感染的样品中成功检测到了这些物种。使用HRM-qPCR分析筛选的87个有袋动物样品中,锥虫为93.1%阳性,而锥虫为8.0%以上。除了三种针对性锥虫物种,该测定法还能够检测和鉴定其他天然和外来锥虫。从样品制备到获得结果,该测定的周转时间少于2小时,对于每种目标锥虫物种,反应的检测限为10拷贝扩增子。这种更快速,更灵敏的诊断工具为锥虫感染的检测,鉴定和定量提供了一个高通量平台。它还将增进对寄主多样性和寄生虫关系的了解,并促进保护管理的决策。


























 京公网安备 11010802027423号
京公网安备 11010802027423号