当前位置:
X-MOL 学术
›
Dev. Growth Differ.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
CRISPR/Cas9 nickase‐mediated efficient and seamless knock‐in of lethal genes in the medaka fish Oryzias latipes
Development, Growth & Differentiation ( IF 2.5 ) Pub Date : 2020-11-05 , DOI: 10.1111/dgd.12700 Yu Murakami 1 , Ryota Futamata 2 , Tomohisa Horibe 3 , Kazumitsu Ueda 4 , Masato Kinoshita 1
Development, Growth & Differentiation ( IF 2.5 ) Pub Date : 2020-11-05 , DOI: 10.1111/dgd.12700 Yu Murakami 1 , Ryota Futamata 2 , Tomohisa Horibe 3 , Kazumitsu Ueda 4 , Masato Kinoshita 1
Affiliation
|
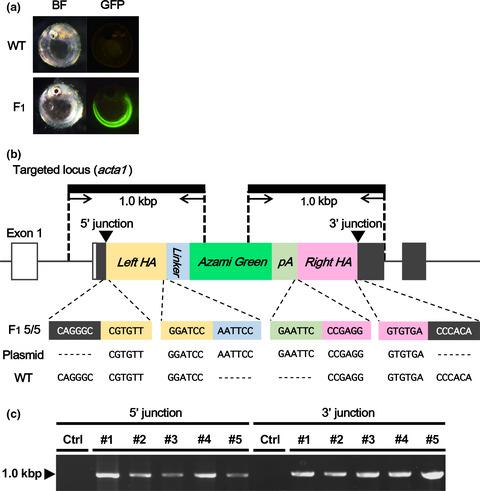
The CRISPR/Cas system offers new opportunities for targeted gene modifications in a wide range of organisms. In medaka (Oryzias latipes), a vertebrate model organism, a wild‐type Cas9‐based approach is commonly used to establish desired strains, however, its use in lethal genes is still challenging due to excess gene disruptions triggered by DNA double strand breaks (DSBs). To overcome this problem, we aimed to develop a new knock‐in system using Cas9 nickase (Cas9n) that can reduce DNA DSBs. We revealed that Cas9n allowed reduction of the DSB‐induced unwanted mutagenesis via non‐homologous end‐joining at both on‐ and off‐ target sites. Further, with a new donor plasmid (p2BaitD) that provides a linear template through Cas9n‐mediated nicks, we successfully integrated reporter cassettes via homology‐directed repair (HDR) into all three loci tested, including a lethal gene. In the experiment targeting the lethal gene, the combination of p2BaitD and Cas9n achieved higher survival rates than the Cas9‐based approach, which enabled the desired knock‐in founders. Additionally, through a technical blend of our knock‐in system with a recently developed One‐step mating protocol, we successfully established a homozygous knock‐in strain in one generation period. This study presents evidence of an effective method to generate an HDR‐mediated gene knock‐in in medaka and other organisms, which is useful for establishing screening platforms for genes or drugs toxicity or other applications.
中文翻译:

CRISPR/Cas9 切口酶介导的青鳉致死基因的高效无缝敲入
CRISPR/Cas 系统为广泛生物体中的靶向基因修饰提供了新的机会。在青鳉 ( Oryzias latipes),一种脊椎动物模型生物,基于野生型 Cas9 的方法通常用于建立所需的菌株,然而,由于 DNA 双链断裂 (DSB) 引发的过量基因破坏,其在致死基因中的应用仍然具有挑战性。为了克服这个问题,我们旨在开发一种使用 Cas9 切口酶 (Cas9n) 的新敲入系统,该系统可以减少 DNA DSB。我们发现 Cas9n 允许通过在目标上和脱靶位点的非同源末端连接减少 DSB 诱导的不需要的诱变。此外,使用通过 Cas9n 介导的切口提供线性模板的新供体质粒 (p2BaitD),我们成功地通过同源定向修复 (HDR) 将报告基因盒整合到所有测试的三个基因座中,包括一个致死基因。在针对致死基因的实验中,p2BaitD 和 Cas9n 的组合比基于 Cas9 的方法获得了更高的存活率,这使得所需的敲入创始人成为可能。此外,通过我们的敲入系统与最近开发的一步交配方案的技术融合,我们成功地在一代期内建立了纯合敲入菌株。本研究提供了在青鳉和其他生物体中产生 HDR 介导的基因敲入的有效方法的证据,这有助于建立基因或药物毒性或其他应用的筛选平台。
更新日期:2020-12-21
中文翻译:

CRISPR/Cas9 切口酶介导的青鳉致死基因的高效无缝敲入
CRISPR/Cas 系统为广泛生物体中的靶向基因修饰提供了新的机会。在青鳉 ( Oryzias latipes),一种脊椎动物模型生物,基于野生型 Cas9 的方法通常用于建立所需的菌株,然而,由于 DNA 双链断裂 (DSB) 引发的过量基因破坏,其在致死基因中的应用仍然具有挑战性。为了克服这个问题,我们旨在开发一种使用 Cas9 切口酶 (Cas9n) 的新敲入系统,该系统可以减少 DNA DSB。我们发现 Cas9n 允许通过在目标上和脱靶位点的非同源末端连接减少 DSB 诱导的不需要的诱变。此外,使用通过 Cas9n 介导的切口提供线性模板的新供体质粒 (p2BaitD),我们成功地通过同源定向修复 (HDR) 将报告基因盒整合到所有测试的三个基因座中,包括一个致死基因。在针对致死基因的实验中,p2BaitD 和 Cas9n 的组合比基于 Cas9 的方法获得了更高的存活率,这使得所需的敲入创始人成为可能。此外,通过我们的敲入系统与最近开发的一步交配方案的技术融合,我们成功地在一代期内建立了纯合敲入菌株。本研究提供了在青鳉和其他生物体中产生 HDR 介导的基因敲入的有效方法的证据,这有助于建立基因或药物毒性或其他应用的筛选平台。



























 京公网安备 11010802027423号
京公网安备 11010802027423号