当前位置:
X-MOL 学术
›
ACS Catal.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Structure and Dynamics of the Reactive State for the Histidine Methylation Process and Catalytic Mechanism of SETD3: Insights from Quantum Mechanics/Molecular Mechanics Investigation
ACS Catalysis ( IF 12.9 ) Pub Date : 2020-11-02 , DOI: 10.1021/acscatal.0c03390 Hao Deng 1 , Yue Ma 2, 3 , Wan-Sheng Ren 1 , Van Quan Vuong 4 , Ping Qian 1 , Hong Guo 2, 3
ACS Catalysis ( IF 12.9 ) Pub Date : 2020-11-02 , DOI: 10.1021/acscatal.0c03390 Hao Deng 1 , Yue Ma 2, 3 , Wan-Sheng Ren 1 , Van Quan Vuong 4 , Ping Qian 1 , Hong Guo 2, 3
Affiliation
|
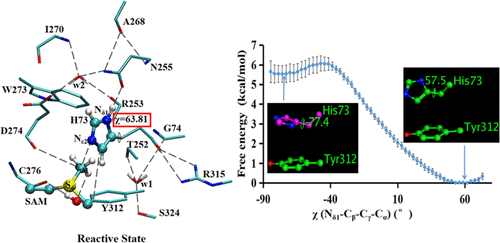
The SETD3 enzyme adds a methyl group to Nε2 of His73 in β-actin, and such methylation finetunes actin’s biochemical properties and cellular function. Here, quantum mechanics/molecular mechanics molecular dynamics and free energy (potential of mean force, PMF) simulations are performed to understand structural and dynamic properties of the reactive state of the SETD3–substrate complex for the methylation process and study the catalytic mechanism of the SETD3 methyltransferase. It is demonstrated that the reactive state for methylation in which His73 adopts the Nδ1–H π tautomeric form is stable and His73 is well positioned at the SETD3 active site for accepting the methyl group from S-adenosyl-l-methionine (SAM). Moreover, the imidazole ring of His73 in the reactive state has a similar orientation to methylated His73 (His73Me) in the product complex. The results suggest that the imidazole ring of His73 does not undergo rotation during the methyl transfer, and this suggestion is further supported by the results of the PMF simulations for the rotation around the Cβ–Cγ bond in the reactive state as well as classical MD simulations. The free energy simulations are also performed for the methyl transfer from SAM to Nε2 of the Nδ1–H π tautomer of His73 in wild-type SETD3 as well as in Asn255Ala and Tyr312Phe mutants. It is shown that the free energy barrier increases by approximately 2–4 kcal mol–1 as a result of these mutations, consistent with the experimental observations. The implication for the existence of the stable reactive state for methylation with well-positioned His73 in the SETD3–substrate complex is discussed, and the origin of the increase of the free energy barrier as a result of the mutations is proposed.
中文翻译:

组氨酸甲基化过程的反应态结构和动力学以及SETD3的催化机理:来自量子力学/分子力学研究的见解
所述SETD3酶添加甲基到N ε2在β肌动蛋白的His73,并且这样的甲基化finetunes肌动蛋白的生化特性和细胞功能。在这里,进行了量子力学/分子力学的分子动力学和自由能(平均力的势,PMF)模拟,以了解SETD3-底物配合物的甲基化过程的反应态的结构和动力学性质,并研究了其催化机理。 SETD3甲基转移酶。已经证明,对于甲基化反应性状态,其中His73采用N个δ1 -Hπ互变异构形式是稳定的,His73是公位于SETD3活性位点用于接收甲基从小号-adenosyl-升-蛋氨酸(SAM)。此外,处于反应状态的His73的咪唑环具有与产物复合物中的甲基化His73(His73Me)相似的取向。结果表明,His73的咪唑环上的甲基转移过程中不发生旋转,并且该建议被PMF为模拟以C旋转的结果进一步支持了β -C γ中的反应性的状态键以及经典MD模拟。自由能仿真还用于从SAM甲基转移至N执行ε2的N δ1在野生型SETD3以及在Asn255Ala和Tyr312Phe突变体-HπHis73的互变异构体。结果表明,自由能垒增加了大约2-4 kcal mol –1这些突变的结果,与实验观察一致。讨论了在SETD3-底物复合物中存在具有良好定位的His73的甲基化稳定反应态的意义,并提出了突变导致自由能垒增加的起源。
更新日期:2020-11-21
中文翻译:

组氨酸甲基化过程的反应态结构和动力学以及SETD3的催化机理:来自量子力学/分子力学研究的见解
所述SETD3酶添加甲基到N ε2在β肌动蛋白的His73,并且这样的甲基化finetunes肌动蛋白的生化特性和细胞功能。在这里,进行了量子力学/分子力学的分子动力学和自由能(平均力的势,PMF)模拟,以了解SETD3-底物配合物的甲基化过程的反应态的结构和动力学性质,并研究了其催化机理。 SETD3甲基转移酶。已经证明,对于甲基化反应性状态,其中His73采用N个δ1 -Hπ互变异构形式是稳定的,His73是公位于SETD3活性位点用于接收甲基从小号-adenosyl-升-蛋氨酸(SAM)。此外,处于反应状态的His73的咪唑环具有与产物复合物中的甲基化His73(His73Me)相似的取向。结果表明,His73的咪唑环上的甲基转移过程中不发生旋转,并且该建议被PMF为模拟以C旋转的结果进一步支持了β -C γ中的反应性的状态键以及经典MD模拟。自由能仿真还用于从SAM甲基转移至N执行ε2的N δ1在野生型SETD3以及在Asn255Ala和Tyr312Phe突变体-HπHis73的互变异构体。结果表明,自由能垒增加了大约2-4 kcal mol –1这些突变的结果,与实验观察一致。讨论了在SETD3-底物复合物中存在具有良好定位的His73的甲基化稳定反应态的意义,并提出了突变导致自由能垒增加的起源。


























 京公网安备 11010802027423号
京公网安备 11010802027423号