Letters in Drug Design & Discovery ( IF 1 ) Pub Date : 2020-10-31 , DOI: 10.2174/1570180817999200630124920 Jian-Bo Tong 1 , Feng Yi 1 , Ding Luo 1 , Tian-Hao Wang 1
|
Background: In recent years, cancer has become the main cause of death and it is a serious threat to human health, so the development of new, selective and safe anticancer drugs is still the focus of medical research. Matrix metalloproteinases-2 (MMP-2) has been determined to play an important role in the regulation of tumor angiogenesis, which is closely related to the development of the tumor. Therefore, MMP-2 is considered as a promising target for tumor therapy. In this study, Tomper comparative molecular field analysis (Topomer CoMFA) and molecular docking were used to investigate the important role of sulfonamide hydroxamate derivatives, an inhibitor of MMP-2, in the inhibition of angiogenesis.
Methods: Quantitative structure active relationship (QSAR) models of 35 sulfonamide hydroxamate derivatives with inhibitory MMPs were developed. The quantitative structure-activity relationship (QSAR) model was built by using Topomer comparative molecular field analysis (Topomer CoMFA) technique.
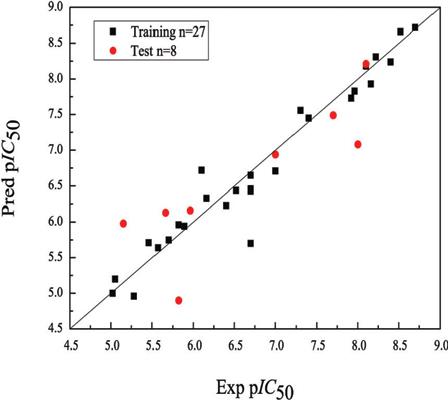
Results and Discussions: The results show that the cross-validated q2 value of the Topomer CoMFA model is 0.881 and the non-cross-validated r2 value is 0.967. The results show that the model is reasonable and reliable, and has good prediction ability. Molecular docking studies were used to find the actual conformations of chemicals in active sites of cancer protease, as well as the binding mode pattern to the binding site in MMP-2. The information provided by the 3D-QSAR model and molecular docking may lead to a better understanding of the structural requirements of 35 sulfonamide hydroxamate derivatives and help to design potential anti-cancer protease inhibitor molecules.
Conclusion: Thirty-five analogs were used in the 3D-QSAR study. Topomer CoMFA 3D-QSAR method was used to build the model, and the model was well predicted and statistically validated. The results of 3D-QSAR and molecular docking analysis provide theoretical guidance for the synthesis of new MMP-2 inhibitors.
中文翻译:

磺酰胺异羟肟酸酯衍生物作为MMP-2抑制剂拓扑CoMFA的QSAR研究和分子对接
背景:近年来,癌症已成为主要的死亡原因,对人类健康构成严重威胁,因此,开发新的,选择性的和安全的抗癌药物仍然是医学研究的重点。已确定基质金属蛋白酶2(MMP-2)在调节肿瘤血管生成中起重要作用,这与肿瘤的发展密切相关。因此,MMP-2被认为是肿瘤治疗的有希望的靶标。在这项研究中,Tomper比较分子场分析(Topomer CoMFA)和分子对接用于研究磺胺异羟肟酸酯衍生物(一种MMP-2抑制剂)在抑制血管生成中的重要作用。
方法:建立了具有抑制性MMPs的35种磺酰胺异羟肟酸酯衍生物的定量构效关系(QSAR)模型。利用拓扑异构体比较分子场分析(Topomer CoMFA)技术建立了定量构效关系(QSAR)模型。
结果与讨论:结果表明,Topomer CoMFA模型的交叉验证的q2值为0.881,未交叉验证的r2值为0.967。结果表明,该模型合理可靠,具有良好的预测能力。分子对接研究用于发现癌症蛋白酶活性位点中化学物质的实际构象,以及与MMP-2中结合位点的结合模式。3D-QSAR模型和分子对接所提供的信息可能有助于更好地理解35种磺酰胺异羟肟酸酯衍生物的结构要求,并有助于设计潜在的抗癌蛋白酶抑制剂分子。
结论:在3D-QSAR研究中使用了35个类似物。使用Topomer CoMFA 3D-QSAR方法构建模型,并对模型进行了良好的预测和统计验证。3D-QSAR和分子对接分析的结果为合成新型MMP-2抑制剂提供了理论指导。

























 京公网安备 11010802027423号
京公网安备 11010802027423号