Journal of Molecular Graphics and Modelling ( IF 2.9 ) Pub Date : 2020-10-07 , DOI: 10.1016/j.jmgm.2020.107770 Niranjan Kumar 1 , Rakesh Srivastava 1 , Amresh Prakash 2 , Andrew M Lynn 1
|
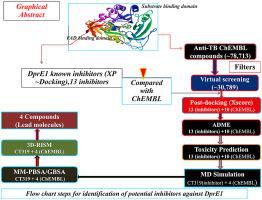
In Mycobacterium tuberculosis (MTB), the cell wall synthesis flavoenzyme decaprenylphosphoryl-β-d-ribose 2′-epimerase (DprE1) plays a crucial role in host pathogenesis, virulence, lethality and survival under stress. The emergence of different variants of drug resistant MTB are a major threat worldwide which essentially requires more effective new drug molecules with no major side effects. Here, we used structure based virtual screening of bioactive molecules from the ChEMBL database targeting DprE1, having bioactive 78,713 molecules known for anti-tuberculosis activity. An extensive molecular docking, binding affinity and pharmacokinetics profile filtering results in the selection four compounds, C5 (ChEMBL2441313), C6 (ChEMBL2338605), C8 (ChEMBL441373) and C10 (ChEMBL1607606) which may explore as potential drug candidates. The obtained results were validated with thirteen known DprE1 inhibitors. We further estimated the free-binding energy, solvation and entropy terms underlying the binding properties of DprE1-ligand interactions with the implication of MD simulation, MM/GBSA, MM/PBSA and MM/3D-RISM. Interestingly, we find that C6 shows the highest ΔG scores (−41.28 ± 3.51, −22.36 ± 3.17, −10.33 ± 5.70 kcal mol−1) in MM/GBSA, MM/PBSA and MM/3D-RISM assay, respectively. Whereas, the lowest ΔG scores (−35.31 ± 3.44, −13.67 ± 2.65, −3.40 ± 4.06 kcal mol−1) observed for CT319, the inhibitor co-crystallized with DprE1. Collectively, the results demonstrated that hit-molecules: C5, C6, C8 and C10 having better binding free energy and molecular affinity as compared to CT319. Thus, we proposed that selected compounds may be explored as lead molecules in MTB therapy.
中文翻译:

虚拟筛选和自由能估计用于鉴定结核分枝杆菌黄酶DprE1抑制剂
在结核分枝杆菌(MTB)中,细胞壁合成黄素酶癸烯酰基磷酸基-β - d-核糖2'-表异构酶(DprE1)在宿主的发病机理,毒力,致死性和应激条件下的存活中起着至关重要的作用。耐药性MTB的不同变体的出现是世界范围内的主要威胁,它本质上需要更有效的新药物分子且没有重大副作用。在这里,我们从ChEMBL数据库中针对DprE1的生物活性分子进行了基于结构的虚拟筛选,该生物活性分子具有抗结核活性已知的78,713个分子。广泛的分子对接,结合亲和力和药代动力学概况过滤导致选择四种化合物C5(ChEMBL2441313),C6(ChEMBL2338605),C8(ChEMBL441373)和C10(ChEMBL1607606),可能会作为潜在的候选药物进行探索。使用十三种已知的DprE1抑制剂验证了获得的结果。我们进一步估计了DprE1-配体相互作用的结合特性背后的自由结合能,溶剂化和熵项,这涉及MD模拟,MM / GBSA,MM / PBSA和MM / 3D-RISM。有趣的是,我们发现在MM / GBSA,MM / PBSA和MM / 3D-RISM分析中,C6分别显示出最高的ΔG得分(−41.28±3.51,−22.36±3.17,−10.33±5.70 kcal mol -1)。而抑制剂CT319的最低ΔG分数(-35.31±3.44,-13.67±2.65,-3.40±4.06 kcal mol -1)最低,该抑制剂与DprE1共结晶。总的来说,结果表明命中分子:与CT319相比,C5,C6,C8和C10具有更好的结合自由能和分子亲和力。因此,我们建议可以将选定的化合物作为MTB治疗中的先导分子进行研究。


























 京公网安备 11010802027423号
京公网安备 11010802027423号