Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Identification of methicillin-resistant Staphylococcus aureus bacteria using surface-enhanced Raman spectroscopy and machine learning techniques
Analyst ( IF 4.2 ) Pub Date : 2020-10-06 , DOI: 10.1039/d0an00476f Fatma Uysal Ciloglu 1, 2, 3, 4 , Ayse Mine Saridag 4, 5, 6, 7 , Ibrahim Halil Kilic 4, 6, 7, 8 , Mahmut Tokmakci 1, 2, 3, 4 , Mehmet Kahraman 4, 5, 6, 7 , Omer Aydin 1, 2, 3, 4, 9
Analyst ( IF 4.2 ) Pub Date : 2020-10-06 , DOI: 10.1039/d0an00476f Fatma Uysal Ciloglu 1, 2, 3, 4 , Ayse Mine Saridag 4, 5, 6, 7 , Ibrahim Halil Kilic 4, 6, 7, 8 , Mahmut Tokmakci 1, 2, 3, 4 , Mehmet Kahraman 4, 5, 6, 7 , Omer Aydin 1, 2, 3, 4, 9
Affiliation
|
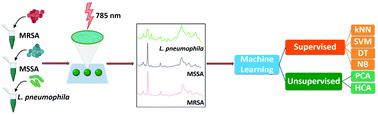
To combat antibiotic resistance, it is extremely important to select the right antibiotic by performing rapid diagnosis of pathogens. Traditional techniques require complicated sample preparation and time-consuming processes which are not suitable for rapid diagnosis. To address this problem, we used surface-enhanced Raman spectroscopy combined with machine learning techniques for rapid identification of methicillin-resistant and methicillin-sensitive Gram-positive Staphylococcus aureus strains and Gram-negative Legionella pneumophila (control group). A total of 10 methicillin-resistant S. aureus (MRSA), 3 methicillin-sensitive S. aureus (MSSA) and 6 L. pneumophila isolates were used. The obtained spectra indicated high reproducibility and repeatability with a high signal to noise ratio. Principal component analysis (PCA), hierarchical cluster analysis (HCA), and various supervised classification algorithms were used to discriminate both S. aureus strains and L. pneumophila. Although there were no noteworthy differences between MRSA and MSSA spectra when viewed with the naked eye, some peak intensity ratios such as 732/958, 732/1333, and 732/1450 proved that there could be a significant indicator showing the difference between them. The k-nearest neighbors (kNN) classification algorithm showed superior classification performance with 97.8% accuracy among the traditional classifiers including support vector machine (SVM), decision tree (DT), and naïve Bayes (NB). Our results indicate that SERS combined with machine learning can be used for the detection of antibiotic-resistant and susceptible bacteria and this technique is a very promising tool for clinical applications.
中文翻译:

使用表面增强拉曼光谱和机器学习技术鉴定耐甲氧西林的金黄色葡萄球菌细菌
为了对抗抗生素耐药性,通过对病原体进行快速诊断来选择正确的抗生素非常重要。传统技术需要复杂的样品制备和耗时的过程,不适合快速诊断。为了解决这个问题,我们使用表面增强拉曼光谱结合机器学习技术来快速鉴定耐甲氧西林和对甲氧西林敏感的革兰氏阳性金黄色葡萄球菌菌株和革兰氏阴性嗜肺军团菌(对照组)。共有10个耐甲氧西林的金黄色葡萄球菌(MRSA),甲氧西林3敏感的金黄色葡萄球菌(MSSA)和6嗜肺军团菌使用分离株。所获得的光谱表明具有高信噪比的高重现性和可重复性。主成分分析(PCA),层次聚类分析(HCA)和各种监督分类算法用于区分金黄色葡萄球菌菌株和嗜肺乳杆菌。尽管用肉眼观察时,MRSA和MSSA光谱之间没有显着差异,但一些峰强度比(例如732 / 958、732 / 1333和732/1450)证明可能存在显着的指标,表明它们之间的差异。该ķ最近邻(kNN)分类算法在支持向量机(SVM),决策树(DT)和朴素贝叶斯(NB)等传统分类器中显示出卓越的分类性能,准确率达97.8%。我们的结果表明,SERS与机器学习相结合可用于检测抗药性和易感细菌,并且该技术是临床应用中非常有前途的工具。
更新日期:2020-11-03
中文翻译:

使用表面增强拉曼光谱和机器学习技术鉴定耐甲氧西林的金黄色葡萄球菌细菌
为了对抗抗生素耐药性,通过对病原体进行快速诊断来选择正确的抗生素非常重要。传统技术需要复杂的样品制备和耗时的过程,不适合快速诊断。为了解决这个问题,我们使用表面增强拉曼光谱结合机器学习技术来快速鉴定耐甲氧西林和对甲氧西林敏感的革兰氏阳性金黄色葡萄球菌菌株和革兰氏阴性嗜肺军团菌(对照组)。共有10个耐甲氧西林的金黄色葡萄球菌(MRSA),甲氧西林3敏感的金黄色葡萄球菌(MSSA)和6嗜肺军团菌使用分离株。所获得的光谱表明具有高信噪比的高重现性和可重复性。主成分分析(PCA),层次聚类分析(HCA)和各种监督分类算法用于区分金黄色葡萄球菌菌株和嗜肺乳杆菌。尽管用肉眼观察时,MRSA和MSSA光谱之间没有显着差异,但一些峰强度比(例如732 / 958、732 / 1333和732/1450)证明可能存在显着的指标,表明它们之间的差异。该ķ最近邻(kNN)分类算法在支持向量机(SVM),决策树(DT)和朴素贝叶斯(NB)等传统分类器中显示出卓越的分类性能,准确率达97.8%。我们的结果表明,SERS与机器学习相结合可用于检测抗药性和易感细菌,并且该技术是临床应用中非常有前途的工具。


























 京公网安备 11010802027423号
京公网安备 11010802027423号