Journal of Proteomics ( IF 3.3 ) Pub Date : 2020-10-02 , DOI: 10.1016/j.jprot.2020.103995 Yu-Na Chai 1 , Jin Qin 2 , Yan-le Li 3 , Ya-Lin Tong 3 , Guang-Hui Liu 4 , Xin-Ru Wang 1 , Cheng-Ye Liu 5 , Ming-Hang Peng 6 , Chong-Zhen Qin 1 , Yu-Rong Xing 7
|
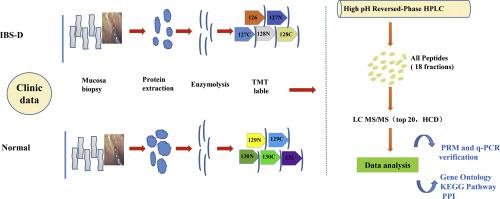
Diarrheal irritable bowel syndrome (IBS-D) is a chronic functional bowel disease with no clear diagnostic markers and no satisfactory treatment strategies. In recent years, the importance of intestinal microstructure and function in IBS-D has been emphasized. However, the intestinal tissue proteomics of IBS-D patients has not been analyzed. Here, we systematically analyzed the molecule profiling of the intestinal tissues in IBS-D patients through tandem mass tag (TMT)-based proteomics for the first time, aiming to reveal the pathogenesis and provide evidence for diagnosis and treatment of IBS-D. Five IBS-D patients and five healthy subjects were selected, biopsy tissue samples from the junction of sigmoid and rectum were analyzed by TMT proteomics. Differentially expressed proteins were obtained and bioinformatics analysis was performed. Furthermore, parallel reaction monitoring (PRM) and q-PCR detection were applied to validate the differentially expressed proteins. Eighty differentially expressed proteins were screened, 48 of which were up-regulated and 32 were down-regulated (fold change >1.2, P < 0.05). Bioinformatics analysis showed that these proteins were significantly enriched in the nutrient ingestion pathways which are related to immune molecules. SELENBP1, VSIG2, HMGB1, DHCR7, BCAP31 and other molecules were significantly changed. Our study revealed the underlying mechanisms of IBS-D intestinal dysfunction.
Significance
Irritable bowel syndrome with diarrhea (IBS-D) is a worldwide chronic intestinal disease with no definite diagnostic markers. It is still a challenge to accurately locate the pathogenesis of patients for appropriate treatment strategy. Established proteomics studies of IBS-D are only based on urine, blood, or tissue samples from animals. Our study was the first TMT proteomics analysis on intestinal biopsy tissues of patients with IBS-D, which revealed the changes of molecular spectrum of actual intestinal conditions in patients with IBS-D. Some important molecules and signaling pathways have been found abnormal in our study, which were related with nutrient uptake. They not only provided preliminary clues for low fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAP) intolerance, an unsolved conundrum of IBS-D, but also revealed obscure problems of protein, lipid, and other nutrients ingestion in IBS-D patients. Some of these differentially expressed molecules have been preliminarily verified, and will may be potential candidate molecules for diagnostic markers of IBS-D.
中文翻译:

肠易激综合征腹泻患者肠道组织的 TMT 蛋白质组学分析:对多种营养摄入异常的影响
腹泻性肠易激综合征(IBS-D)是一种慢性功能性肠病,没有明确的诊断标志物,也没有令人满意的治疗策略。近年来,肠道微结构和功能在 IBS-D 中的重要性得到了强调。然而,尚未分析 IBS-D 患者的肠组织蛋白质组学。在这里,我们首次通过基于串联质量标签(TMT)的蛋白质组学系统分析了IBS-D患者肠道组织的分子谱,旨在揭示发病机制,为IBS-D的诊断和治疗提供依据。选取 5 名 IBS-D 患者和 5 名健康受试者,对乙状结肠和直肠交界处的活检组织样本进行 TMT 蛋白质组学分析。获得差异表达的蛋白质并进行生物信息学分析。此外,应用平行反应监测 (PRM) 和 q-PCR 检测来验证差异表达的蛋白质。筛选出80个差异表达蛋白,其中48个上调,32个下调(倍数变化>1.2,P < 0.05)。生物信息学分析表明,这些蛋白质在与免疫分子相关的营养摄取途径中显着富集。SELENBP1、VSIG2、HMGB1、DHCR7、BCAP31等分子发生显着变化。我们的研究揭示了 IBS-D 肠功能障碍的潜在机制。
意义
伴有腹泻的肠易激综合征 (IBS-D) 是一种全球性的慢性肠道疾病,没有明确的诊断标志物。准确定位患者的发病机制以制定合适的治疗策略仍然是一个挑战。已建立的 IBS-D 蛋白质组学研究仅基于来自动物的尿液、血液或组织样本。我们的研究是首次对 IBS-D 患者肠道活检组织进行 TMT 蛋白质组学分析,揭示了 IBS-D 患者实际肠道状况的分子谱变化。在我们的研究中发现一些重要的分子和信号通路异常,与营养吸收有关。他们不仅为低发酵寡糖、二糖、单糖和多元醇 (FODMAP) 不耐受提供了初步线索,这是 IBS-D 的一个未解之谜,但也揭示了 IBS-D 患者摄入蛋白质、脂质和其他营养物质的模糊问题。其中一些差异表达分子已得到初步验证,可能成为IBS-D诊断标志物的潜在候选分子。


























 京公网安备 11010802027423号
京公网安备 11010802027423号