当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
PulseDIA: Data-Independent Acquisition Mass Spectrometry Using Multi-Injection Pulsed Gas-Phase Fractionation
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2020-09-25 , DOI: 10.1021/acs.jproteome.0c00381 Xue Cai 1, 2 , Weigang Ge 1, 2 , Xiao Yi 1, 2 , Rui Sun 1, 2 , Jiang Zhu 3 , Cong Lu 3 , Ping Sun 4 , Tiansheng Zhu 1, 2 , Guan Ruan 1, 2 , Chunhui Yuan 1, 2 , Shuang Liang 1, 2 , Mengge Lyu 1, 2 , Shiang Huang 3 , Yi Zhu 1, 2 , Tiannan Guo 1, 2
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2020-09-25 , DOI: 10.1021/acs.jproteome.0c00381 Xue Cai 1, 2 , Weigang Ge 1, 2 , Xiao Yi 1, 2 , Rui Sun 1, 2 , Jiang Zhu 3 , Cong Lu 3 , Ping Sun 4 , Tiansheng Zhu 1, 2 , Guan Ruan 1, 2 , Chunhui Yuan 1, 2 , Shuang Liang 1, 2 , Mengge Lyu 1, 2 , Shiang Huang 3 , Yi Zhu 1, 2 , Tiannan Guo 1, 2
Affiliation
|
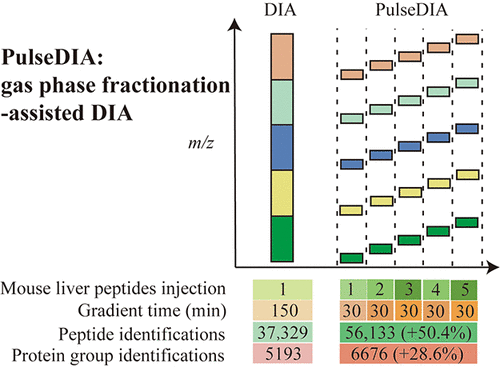
The performance of data-independent acquisition (DIA) mass spectrometry (MS) depends on the separation efficiency of peptide precursors. In Orbitrap-based mass spectrometers, separation efficiency of peptide precursors is limited by the relatively slow scanning rate compared to time of flight (TOF)-based MS. Here, we present PulseDIA, a multi-injection gas-phase fractionation (GPF) strategy for enhanced DIA-MS. This is achieved by equally dividing the conventional DIA analysis covering the entire mass range into multiple injections for DIA analyses with complementary windows. Using mouse liver digests, the PulseDIA method identified up to 50% more peptides and 29% more protein groups than that by conventional DIA with the same length of effective gradient time. Compared to conventional multi-injection GPF, PusleDIA exhibited higher flexibility and identified up to 18% more peptides and 8% more protein groups using two injections. The gain of peptides per effective time unit was the highest in PulseDIA compared to conventional DIA and GPF. We further applied the PulseDIA method to profile the proteome of 18 human tissue samples (benign and malignant) from nine cholangiocarcinoma (CCA) patients. PulseDIA identified 7796 protein groups in these CCA samples, with a 14% increase of protein group identification compared to the conventional DIA method. The missing value for protein matrix dropped by 7% using PulseDIA compared to DIA. A total of 681 significantly altered proteins were detected in CCA samples using PulseDIA, including several dysregulated proteins, which were absent in the conventional DIA analysis. Taken together, we present PulseDIA as an enhanced DIA-MS method with improved sensitivity and reproducibility.
中文翻译:

PulseDIA:使用多进样脉冲气相色谱分离技术的数据独立采集质谱
数据独立采集(DIA)质谱(MS)的性能取决于肽前体的分离效率。在基于Orbitrap的质谱仪中,与基于飞行时间(TOF)的MS相比,相对较慢的扫描速率限制了肽前体的分离效率。在这里,我们介绍PulseDIA,这是一种用于增强DIA-MS的多进样气相分级(GPF)策略。这是通过将覆盖整个质量范围的常规DIA分析平均分为多个进样来进行的,用于带有互补窗口的DIA分析。使用小鼠肝脏消化物,PulseDIA方法比具有相同有效梯度时间长度的常规DIA所鉴定的肽多出50%,肽和蛋白质组多29%。与传统的多次进样GPF相比,PusleDIA具有更高的灵活性,并且使用两次进样可鉴定出多达18%的肽和8%的蛋白质组。与传统的DIA和GPF相比,PulseDIA中每有效时间单位的肽增益最高。我们进一步应用PulseDIA方法对来自9位胆管癌(CCA)患者的18个人体组织样品(良性和恶性)进行蛋白质组分析。PulseDIA在这些CCA样品中鉴定出7796个蛋白质组,与常规DIA方法相比,蛋白质组鉴定增加了14%。与DIA相比,使用PulseDIA可以使蛋白质基质的缺失值降低7%。使用PulseDIA在CCA样品中总共检测到681种显着改变的蛋白质,其中包括常规DIA分析中不存在的几种失调的蛋白质。在一起
更新日期:2020-09-25
中文翻译:

PulseDIA:使用多进样脉冲气相色谱分离技术的数据独立采集质谱
数据独立采集(DIA)质谱(MS)的性能取决于肽前体的分离效率。在基于Orbitrap的质谱仪中,与基于飞行时间(TOF)的MS相比,相对较慢的扫描速率限制了肽前体的分离效率。在这里,我们介绍PulseDIA,这是一种用于增强DIA-MS的多进样气相分级(GPF)策略。这是通过将覆盖整个质量范围的常规DIA分析平均分为多个进样来进行的,用于带有互补窗口的DIA分析。使用小鼠肝脏消化物,PulseDIA方法比具有相同有效梯度时间长度的常规DIA所鉴定的肽多出50%,肽和蛋白质组多29%。与传统的多次进样GPF相比,PusleDIA具有更高的灵活性,并且使用两次进样可鉴定出多达18%的肽和8%的蛋白质组。与传统的DIA和GPF相比,PulseDIA中每有效时间单位的肽增益最高。我们进一步应用PulseDIA方法对来自9位胆管癌(CCA)患者的18个人体组织样品(良性和恶性)进行蛋白质组分析。PulseDIA在这些CCA样品中鉴定出7796个蛋白质组,与常规DIA方法相比,蛋白质组鉴定增加了14%。与DIA相比,使用PulseDIA可以使蛋白质基质的缺失值降低7%。使用PulseDIA在CCA样品中总共检测到681种显着改变的蛋白质,其中包括常规DIA分析中不存在的几种失调的蛋白质。在一起



























 京公网安备 11010802027423号
京公网安备 11010802027423号