Current Bioinformatics ( IF 4 ) Pub Date : 2020-05-31 , DOI: 10.2174/1574893615666191219095817 Fernando Mora-Márquez 1 , José Luis Vázquez-Poletti 2 , Víctor Chano 1 , Carmen Collada 1 , Álvaro Soto 1 , Unai López de Heredia 1
|
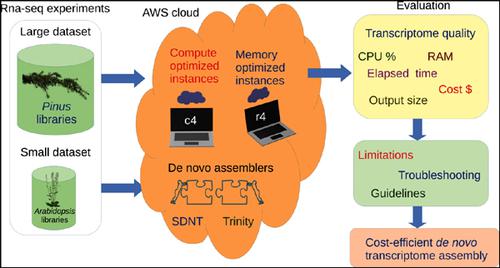
Background: Bioinformatics software for RNA-seq analysis has a high computational requirement in terms of the number of CPUs, RAM size, and processor characteristics. Specifically, de novo transcriptome assembly demands large computational infrastructure due to the massive data size, and complexity of the algorithms employed. Comparative studies on the quality of the transcriptome yielded by de novo assemblers have been previously published, lacking, however, a hardware efficiency-oriented approach to help select the assembly hardware platform in a cost-efficient way.
Objective: We tested the performance of two popular de novo transcriptome assemblers, Trinity and SOAPdenovo-Trans (SDNT), in terms of cost-efficiency and quality to assess limitations, and provided troubleshooting and guidelines to run transcriptome assemblies efficiently.
Methods: We built virtual machines with different hardware characteristics (CPU number, RAM size) in the Amazon Elastic Compute Cloud of the Amazon Web Services. Using simulated and real data sets, we measured the elapsed time, cost, CPU percentage and output size of small and large data set assemblies.
Results: For small data sets, SDNT outperformed Trinity by an order the magnitude, significantly reducing the time duration and costs of the assembly. For large data sets, Trinity performed better than SDNT. Both the assemblers provide good quality transcriptomes.
Conclusion: The selection of the optimal transcriptome assembler and provision of computational resources depend on the combined effect of size and complexity of RNA-seq experiments.
中文翻译:

Amazon Elastic Compute Cloud中的从头转录组组装软件的硬件性能评估
背景:用于RNA序列分析的生物信息学软件对CPU数量,RAM大小和处理器特性有很高的计算要求。具体来说,从头转录组组装需要大量的计算基础结构,这是因为数据量巨大,并且所用算法复杂。以前已经发表了有关从头组装者产生的转录组质量的比较研究,但是缺乏以硬件效率为导向的方法来帮助以经济高效的方式选择组装硬件平台。
目的:我们从成本效率和质量方面评估了局限性,测试了两种流行的从头转录组组装商Trinity和SOAPdenovo-Trans(SDNT)的性能,并提供了故障排除和指导以有效地运行转录组组装。
方法:我们在Amazon Web Services的Amazon Elastic Compute Cloud中构建了具有不同硬件特性(CPU编号,RAM大小)的虚拟机。使用模拟和真实数据集,我们测量了大小数据集程序集的经过时间,成本,CPU百分比和输出大小。
结果:对于小型数据集,SDNT比Trinity高出一个数量级,从而显着减少了安装时间和成本。对于大型数据集,Trinity的性能优于SDNT。两种汇编器均提供高质量的转录组。
结论:最佳转录组组装器的选择和计算资源的提供取决于RNA-seq实验的大小和复杂性的综合影响。


























 京公网安备 11010802027423号
京公网安备 11010802027423号