Current Bioinformatics ( IF 4 ) Pub Date : 2020-06-30 , DOI: 10.2174/1574893614666190902155811 Yunyun Liang 1 , Shengli Zhang 2
|
Background: Apoptosis proteins have a key role in the development and the homeostasis of the organism, and are very important to understand the mechanism of cell proliferation and death. The function of apoptosis protein is closely related to its subcellular location.
Objective: Prediction of apoptosis protein subcellular localization is a meaningful task.
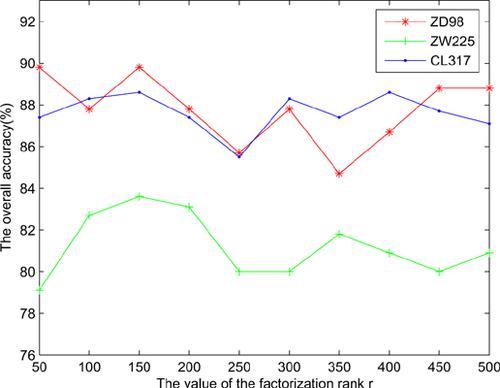
Methods: In this study, we predict the apoptosis protein subcellular location by using the PSSMbased second-order moving average descriptor, nonnegative matrix factorization based on Kullback-Leibler divergence and over-sampling algorithms. This model is named by SOMAPKLNMF- OS and constructed on the ZD98, ZW225 and CL317 benchmark datasets. Then, the support vector machine is adopted as the classifier, and the bias-free jackknife test method is used to evaluate the accuracy.
Results: Our prediction system achieves the favorable and promising performance of the overall accuracy on the three datasets and also outperforms the other listed models.
Conclusion: The results show that our model offers a high throughput tool for the identification of apoptosis protein subcellular localization.
中文翻译:

整合二阶移动平均和过采样算法来预测细胞凋亡蛋白亚细胞定位
背景:凋亡蛋白在生物的发育和体内稳态中起着关键作用,对于理解细胞增殖和死亡的机制非常重要。凋亡蛋白的功能与其亚细胞位置密切相关。
目的:预测凋亡蛋白的亚细胞定位是一项有意义的任务。
方法:在这项研究中,我们通过使用基于PSSM的二阶移动平均描述符,基于Kullback-Leibler散度和过采样算法的非负矩阵分解来预测凋亡蛋白的亚细胞位置。该模型由SOMAPKLNMF- OS命名,并基于ZD98,ZW225和CL317基准数据集构建。然后,采用支持向量机作为分类器,并采用无偏差折刀试验方法进行精度评估。
结果:我们的预测系统在这三个数据集上均取得了总体准确性的良好和有希望的表现,并且也优于其他列出的模型。
结论:结果表明,我们的模型为鉴定凋亡蛋白亚细胞定位提供了高通量的工具。

























 京公网安备 11010802027423号
京公网安备 11010802027423号