当前位置:
X-MOL 学术
›
Biopolymers
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Unnatural bases for recognition of noncoding nucleic acid interfaces
Biopolymers ( IF 2.9 ) Pub Date : 2020-09-24 , DOI: 10.1002/bip.23399 Shiqin Miao 1 , Yufeng Liang 1 , Sarah Rundell 1 , Debmalya Bhunia 1 , Shekar Devari 1 , Oliver Munyaradzi 1 , Dennis Bong 1
Biopolymers ( IF 2.9 ) Pub Date : 2020-09-24 , DOI: 10.1002/bip.23399 Shiqin Miao 1 , Yufeng Liang 1 , Sarah Rundell 1 , Debmalya Bhunia 1 , Shekar Devari 1 , Oliver Munyaradzi 1 , Dennis Bong 1
Affiliation
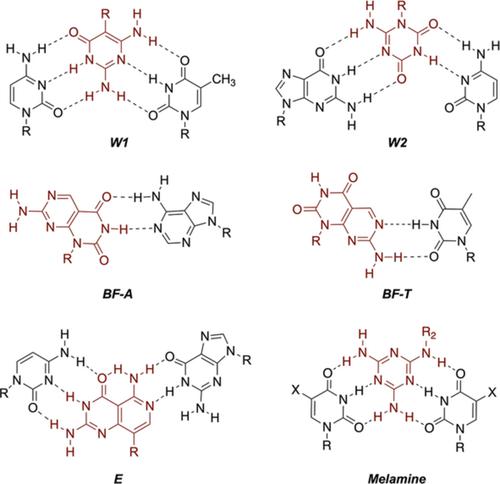
|
The notion of using synthetic heterocycles instead of the native bases to interface with DNA and RNA has been explored for nearly 60 years. Unnatural bases compatible with the DNA/RNA coding interface have the potential to expand the genetic code and co‐opt the machinery of biology to access new macromolecular function; accordingly, this body of research is core to synthetic biology. While much of the literature on artificial bases focuses on code expansion, there is a significant and growing effort on docking synthetic heterocycles to noncoding nucleic acid interfaces; this approach seeks to illuminate major processes of nucleic acids, including regulation of transcription, translation, transport, and transcript lifetimes. These major avenues of research at the coding and noncoding interfaces have in common fundamental principles in molecular recognition. Herein, we provide an overview of foundational literature in biophysics of base recognition and unnatural bases in coding to provide context for the developing area of targeting noncoding nucleic acid interfaces with synthetic bases, with a focus on systems developed through iterative design and biophysical study.
中文翻译:

用于识别非编码核酸界面的非天然碱基
近 60 年来,人们一直在探索使用合成杂环代替天然碱基来连接 DNA 和 RNA 的概念。与 DNA/RNA 编码界面兼容的非天然碱基有可能扩展遗传密码并选择生物学机制来获得新的大分子功能;因此,这一研究主体是合成生物学的核心。虽然许多关于人工碱基的文献都集中在代码扩展上,但在将合成杂环对接到非编码核酸界面方面,人们付出了大量且不断增加的努力。这种方法旨在阐明核酸的主要过程,包括转录、翻译、运输和转录本寿命的调节。编码和非编码接口的这些主要研究途径在分子识别方面具有共同的基本原理。在此,我们概述了碱基识别的生物物理学和编码中的非自然碱基的基础文献,为靶向非编码核酸与合成碱基界面的开发领域提供背景,重点是通过迭代设计和生物物理研究开发的系统。
更新日期:2020-09-24
中文翻译:

用于识别非编码核酸界面的非天然碱基
近 60 年来,人们一直在探索使用合成杂环代替天然碱基来连接 DNA 和 RNA 的概念。与 DNA/RNA 编码界面兼容的非天然碱基有可能扩展遗传密码并选择生物学机制来获得新的大分子功能;因此,这一研究主体是合成生物学的核心。虽然许多关于人工碱基的文献都集中在代码扩展上,但在将合成杂环对接到非编码核酸界面方面,人们付出了大量且不断增加的努力。这种方法旨在阐明核酸的主要过程,包括转录、翻译、运输和转录本寿命的调节。编码和非编码接口的这些主要研究途径在分子识别方面具有共同的基本原理。在此,我们概述了碱基识别的生物物理学和编码中的非自然碱基的基础文献,为靶向非编码核酸与合成碱基界面的开发领域提供背景,重点是通过迭代设计和生物物理研究开发的系统。



























 京公网安备 11010802027423号
京公网安备 11010802027423号