Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Predicting Drug-Induced Liver Injury Using Convolutional Neural Network and Molecular Fingerprint-Embedded Features
ACS Omega ( IF 4.1 ) Pub Date : 2020-09-22 , DOI: 10.1021/acsomega.0c03866 Thanh-Hoang Nguyen-Vo 1 , Loc Nguyen 2 , Nguyet Do 2 , Phuc H. Le 2 , Thien-Ngan Nguyen 2 , Binh P. Nguyen 1 , Ly Le 2, 3
ACS Omega ( IF 4.1 ) Pub Date : 2020-09-22 , DOI: 10.1021/acsomega.0c03866 Thanh-Hoang Nguyen-Vo 1 , Loc Nguyen 2 , Nguyet Do 2 , Phuc H. Le 2 , Thien-Ngan Nguyen 2 , Binh P. Nguyen 1 , Ly Le 2, 3
Affiliation
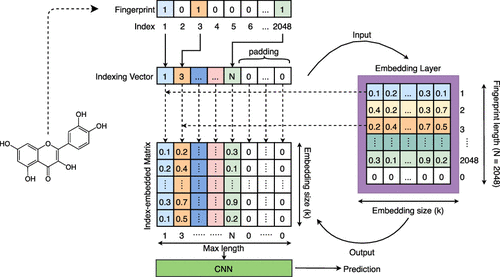
|
As a critical issue in drug development and postmarketing safety surveillance, drug-induced liver injury (DILI) leads to failures in clinical trials as well as retractions of on-market approved drugs. Therefore, it is important to identify DILI compounds in the early-stages through in silico and in vivo studies. It is difficult using conventional safety testing methods, since the predictive power of most of the existing frameworks is insufficiently effective to address this pharmacological issue. In our study, we employ a natural language processing (NLP) inspired computational framework using convolutional neural networks and molecular fingerprint-embedded features. Our development set and independent test set have 1597 and 322 compounds, respectively. These samples were collected from previous studies and matched with established chemical databases for structural validity. Our study comes up with an average accuracy of 0.89, Matthews’s correlation coefficient (MCC) of 0.80, and an AUC of 0.96. Our results show a significant improvement in the AUC values compared to the recent best model with a boost of 6.67%, from 0.90 to 0.96. Also, based on our findings, molecular fingerprint-embedded featurizer is an effective molecular representation for future biological and biochemical studies besides the application of classic molecular fingerprints.
中文翻译:

使用卷积神经网络和分子指纹嵌入功能预测药物诱发的肝损伤
作为药物开发和上市后安全监控的关键问题,药物诱发的肝损伤(DILI)导致临床试验失败以及撤回市场上认可的药物。因此,重要的是通过计算机和体内研究在早期识别DILI化合物。由于大多数现有框架的预测能力不足以有效解决该药理问题,因此使用常规安全性测试方法非常困难。在我们的研究中,我们使用了自然语言处理(NLP)启发的计算框架,该框架使用了卷积神经网络和分子指纹嵌入功能。我们的开发套件和独立测试套件分别包含1597和322种化合物。这些样品是从以前的研究中收集的,并与建立的化学数据库相匹配,以确保结构的有效性。我们的研究得出的平均准确度为0.89,马修斯的相关系数(MCC)为0.80,AUC为0.96。我们的结果表明,与最近的最佳模型相比,AUC值有了显着提高,从0.90到0.96增长了6.67%。另外,根据我们的发现,除经典分子指纹的应用外,分子指纹嵌入的增效剂是未来生物学和生化研究的有效分子代表。我们的结果表明,与最近的最佳模型相比,AUC值有了显着提高,从0.90到0.96增长了6.67%。另外,根据我们的发现,除经典分子指纹的应用外,分子指纹嵌入的增效剂是未来生物学和生化研究的有效分子代表。我们的结果表明,与最近的最佳模型相比,AUC值有了显着提高,从0.90到0.96增长了6.67%。另外,根据我们的发现,除经典分子指纹的应用外,分子指纹嵌入的增效剂是未来生物学和生化研究的有效分子代表。
更新日期:2020-10-06
中文翻译:

使用卷积神经网络和分子指纹嵌入功能预测药物诱发的肝损伤
作为药物开发和上市后安全监控的关键问题,药物诱发的肝损伤(DILI)导致临床试验失败以及撤回市场上认可的药物。因此,重要的是通过计算机和体内研究在早期识别DILI化合物。由于大多数现有框架的预测能力不足以有效解决该药理问题,因此使用常规安全性测试方法非常困难。在我们的研究中,我们使用了自然语言处理(NLP)启发的计算框架,该框架使用了卷积神经网络和分子指纹嵌入功能。我们的开发套件和独立测试套件分别包含1597和322种化合物。这些样品是从以前的研究中收集的,并与建立的化学数据库相匹配,以确保结构的有效性。我们的研究得出的平均准确度为0.89,马修斯的相关系数(MCC)为0.80,AUC为0.96。我们的结果表明,与最近的最佳模型相比,AUC值有了显着提高,从0.90到0.96增长了6.67%。另外,根据我们的发现,除经典分子指纹的应用外,分子指纹嵌入的增效剂是未来生物学和生化研究的有效分子代表。我们的结果表明,与最近的最佳模型相比,AUC值有了显着提高,从0.90到0.96增长了6.67%。另外,根据我们的发现,除经典分子指纹的应用外,分子指纹嵌入的增效剂是未来生物学和生化研究的有效分子代表。我们的结果表明,与最近的最佳模型相比,AUC值有了显着提高,从0.90到0.96增长了6.67%。另外,根据我们的发现,除经典分子指纹的应用外,分子指纹嵌入的增效剂是未来生物学和生化研究的有效分子代表。



























 京公网安备 11010802027423号
京公网安备 11010802027423号