Computational and Structural Biotechnology Journal ( IF 6 ) Pub Date : 2020-09-20 , DOI: 10.1016/j.csbj.2020.09.012 Samuel M. Chekabab , John R. Lawrence , Alvin Alvarado , Bernardo Predicala , Darren R. Korber
|
New Canadian regulations have required that all use of antibiotics in livestock animal production should be under veterinary prescription and oversight, while the prophylactic use and inclusion of these agents in animal feed as growth promoters are also banned. In response to this new rule, many Canadian animal producers have voluntarily implemented production practices aimed at producing animals effectively while avoiding the use of antibiotics. In the swine industry, one such program is the ‘raised without antibiotics’ (RWA) program.
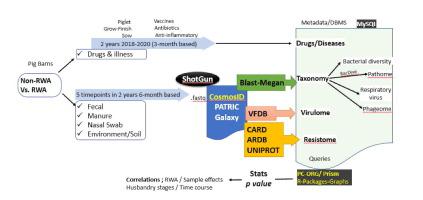
In this paper, we describe a comprehensive investigative methodology comparing the effect of the adoption of the RWA approach with non-RWA pig production operations where antibiotics may still be administered on animals as needed. Our experimental approach involves a multi-year longitudinal investigation of pig farming to determine the effects of antibiotic usage on the prevalence of antimicrobial resistance (AMR) and pathogen abundance in the context of the drug exposures recorded in the RWA versus non-RWA scenarios.
Surveillance of AMR and pathogens was conducted using whole-genome sequencing (WGS) in conjunction with open source tools and data pipeline analyses, which inform on the resistome, virulome and bacterial diversity in animals and materials associated with the different types of barns. This information was combined and correlated with drug usage (types and amounts) over time, along with animal health metadata (stage of growth, reason for drug use, among others). The overarching goal was to develop a set of interconnected informatic tools and data management procedures wherein specific queries could be made and customized, to reveal statistically valid cause/effect relationships.
Results demonstrating possible correlations between RWA and AMR would support the Canadian pig industry, as well as regulatory agencies in new efforts, focused on reducing overall antibiotics use and in curbing the development and spread of AMR related to animal agriculture.
中文翻译:

基于健康元数据的管理方法,用于高通量遗传序列的比较分析,以量化加拿大养猪场的抗菌素耐药性降低
加拿大的新法规要求在牲畜动物生产中所有抗生素的使用均应遵循兽医的规定和监督,同时也禁止将这些药物作为生长促进剂进行预防性使用和包括在动物饲料中。为了响应这一新规定,许多加拿大动物生产者自愿实施了生产实践,旨在有效地生产动物,同时避免使用抗生素。在养猪业中,一项这样的计划是“无抗生素饲养”(RWA)计划。
在本文中,我们描述了一种综合的调查方法,将采用RWA方法与非RWA猪生产操作的效果进行比较,在非RWA猪生产操作中,仍可以根据需要对动物施用抗生素。我们的实验方法涉及对养猪场进行的多年纵向调查,以确定在RWA与非RWA情况下记录的药物暴露情况下,抗生素使用对抗菌素耐药性(AMR)和病原体丰富度的影响。
使用全基因组测序(WGS)结合开源工具和数据流水线分析对AMR和病原体进行监测,从而了解与不同类型的谷仓相关的动物和材料中的抗虫基因组,病毒组和细菌多样性。这些信息经过组合并与一段时间内的药物使用情况(类型和数量)以及动物健康元数据(生长阶段,药物使用原因等)相关联。总体目标是开发一套相互联系的信息工具和数据管理程序,在其中可以进行和定制特定的查询,以揭示统计上有效的因果关系。
证明RWA和AMR之间可能存在相关性的结果将支持加拿大养猪业以及监管机构的新努力,重点是减少整体抗生素的使用并遏制与动物农业有关的AMR的发展和传播。



























 京公网安备 11010802027423号
京公网安备 11010802027423号