Chemosphere ( IF 8.8 ) Pub Date : 2020-09-18 , DOI: 10.1016/j.chemosphere.2020.128331 Meri Nakajima , Reiko Hirano , Satoshi Okabe , Hisashi Satoh
|
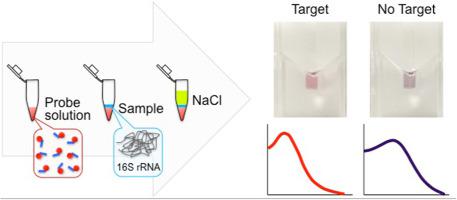
Domestic and industrial wastewater treatment systems are vital in the protection of natural ecosystems and human health. Identification of microbial communities in the systems is essential to stable treatment performance. However, the current tools of microbial community analysis are labor intensive and time consuming, and require expensive equipment. Therefore, we developed a simple assay for colorimetric quantification of bacterial 16S rRNA extracted from environmental samples. The assay is based on RNA extraction with commercial kits, mixing the unamplified RNA sample with Au-nanoprobes and NaCl, and analyzing the absorbance spectra. Our experimental results confirmed that the assay format was valid. By analyzing the synthesized DNA, we optimized the operational parameters affecting the assay. We achieved adequate capture DNA density by setting the capture DNA probe concentration at 10 μM during the functionalization step. The required incubation time after NaCl addition was 30 min. The binding site of the target had negligible effect on DNA detection. Under the optimized condition, a calibration curve was created using 16S rRNA extracted from activated sludge. The curve was linear above 5.0×107 copies/μL of bacterial 16S rRNA concentration, and the limit of detection was 1.17×108 copies/μL. Using the calibration curve, the bacterial 16S rRNA concentration in activated sludge samples could be quantified with deviations between 48% and 208% against those determined by RT-qPCR. The findings of our study introduce an innovative tool for the quantification of 16S rRNA concentration as the activity of key bacteria in wastewater treatment processes, achieving stable treatment performance.
中文翻译:

使用金纳米探针对活性污泥中未扩增细菌16S rRNA进行比色定量的简单测定
家庭和工业废水处理系统对于保护自然生态系统和人类健康至关重要。鉴定系统中的微生物群落对于稳定治疗性能至关重要。然而,当前的微生物群落分析工具劳动强度大且耗时,并且需要昂贵的设备。因此,我们开发了一种简单的测定法,用于比色定量分析从环境样品中提取的细菌16S rRNA。该测定基于使用商业试剂盒的RNA提取,将未扩增的RNA样品与金纳米探针和NaCl混合,并分析吸收光谱。我们的实验结果证实了测定形式是有效的。通过分析合成的DNA,我们优化了影响测定的操作参数。通过在功能化步骤中将捕获DNA探针的浓度设置为10μM,我们获得了足够的捕获DNA密度。加入氯化钠后所需的孵育时间为30分钟。靶标的结合位点对DNA检测的影响可忽略不计。在优化的条件下,使用从活性污泥中提取的16S rRNA绘制校准曲线。曲线在5.0×10以上呈线性细菌16S rRNA浓度为7拷贝/μL,检出限为1.17×10 8拷贝/μL。使用校准曲线,可以定量分析活性污泥样品中细菌16S rRNA的浓度,其相对于RT-qPCR测定的偏差在48%至208%之间。我们研究的结果引入了一种创新的工具,用于量化16S rRNA浓度作为废水处理过程中关键细菌的活性,从而实现稳定的处理性能。



























 京公网安备 11010802027423号
京公网安备 11010802027423号