当前位置:
X-MOL 学术
›
Biotechnol. Bioeng.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Data-Driven In-silico Prediction of Regulation Heterogeneity and ATP Demands of Escherichia coli in Large-scale Bioreactors.
Biotechnology and Bioengineering ( IF 3.8 ) Pub Date : 2020-09-17 , DOI: 10.1002/bit.27568 Julia Zieringer 1 , Moritz Wild 1 , Ralf Takors 1
Biotechnology and Bioengineering ( IF 3.8 ) Pub Date : 2020-09-17 , DOI: 10.1002/bit.27568 Julia Zieringer 1 , Moritz Wild 1 , Ralf Takors 1
Affiliation
|
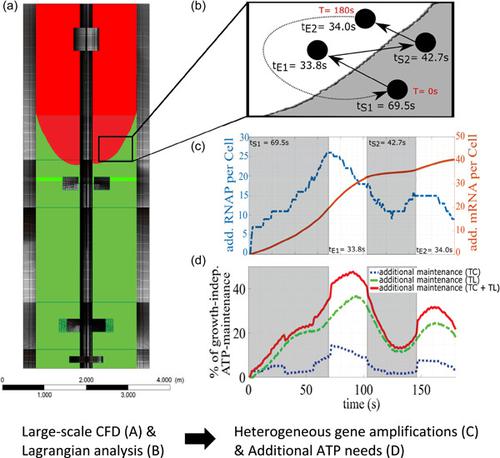
Escherichia coli exposed to industrial‐scale heterogeneous mixing conditions respond to external stress by initiating short‐term metabolic and long‐term strategic transcriptional programs. In native habitats, long‐term strategies allow survival in severe stress but are of limited use in large bioreactors, where microenvironmental conditions may change right after said programs are started. Related on/off switching of genes causes additional ATP burden that may reduce the cellular capacity for producing the desired product. Here, we present an agent‐based data‐driven model linked to computational fluid dynamics, finally allowing to predict additional ATP needs of Escherichia coli K12 W3110 exposed to realistic large‐scale bioreactor conditions. The complex model describes transcriptional up‐ and downregulation dynamics of about 600 genes starting from subminute range covering 28 h. The data‐based approach was extracted from comprehensive scale‐down experiments. Simulating mixing and mass transfer conditions in a 54 m3 stirred bioreactor, 120,000 E. coli cells were tracked while fluctuating between different zones of glucose availability. It was found that cellular ATP demands rise between 30% and 45% of growth decoupled maintenance needs, which may limit the production of ATP‐intensive product formation accordingly. Furthermore, spatial analysis of individual cell transcriptional patterns reveal very heterogeneous gene amplifications with hot spots of 50%–80% messenger RNA upregulation in the upper region of the bioreactor. The phenomenon reflects the time‐delayed regulatory response of the cells that propagate through the stirred tank. After 4.2 h, cells adapt to environmental changes but still have to bear an additional 6% ATP demand.
中文翻译:

大型生物反应器中大肠杆菌调控异质性和 ATP 需求的数据驱动计算机内预测。
暴露于工业规模异质混合条件下的大肠杆菌通过启动短期代谢和长期战略转录程序来响应外部压力。在原生栖息地,长期策略允许在严重压力下生存,但在大型生物反应器中的用途有限,在所述程序启动后微环境条件可能会立即发生变化。相关的基因开/关切换会导致额外的 ATP 负担,这可能会降低细胞生产所需产品的能力。在这里,我们提出了一个与计算流体动力学相关的基于代理的数据驱动模型,最终允许预测大肠杆菌的额外 ATP 需求K12 W3110 暴露于真实的大型生物反应器条件。复杂模型描述了大约 600 个基因的转录上调和下调动态,从覆盖 28 小时的亚分钟范围开始。基于数据的方法是从全面的缩小实验中提取的。在 54 m 3搅拌生物反应器中模拟混合和传质条件,120,000大肠杆菌在不同葡萄糖可用性区域之间波动时跟踪细胞。结果表明,细胞 ATP 需求上升了增长解耦维持需求的 30% 至 45%,这可能会相应地限制 ATP 密集型产品形成的生产。此外,单个细胞转录模式的空间分析揭示了非常异质的基因扩增,在生物反应器的上部区域有 50%–80% 的信使 RNA 上调热点。这种现象反映了通过搅拌罐传播的细胞的延时调节反应。4.2 小时后,细胞适应环境变化,但仍需承受额外的 6% ATP 需求。
更新日期:2020-09-17
中文翻译:

大型生物反应器中大肠杆菌调控异质性和 ATP 需求的数据驱动计算机内预测。
暴露于工业规模异质混合条件下的大肠杆菌通过启动短期代谢和长期战略转录程序来响应外部压力。在原生栖息地,长期策略允许在严重压力下生存,但在大型生物反应器中的用途有限,在所述程序启动后微环境条件可能会立即发生变化。相关的基因开/关切换会导致额外的 ATP 负担,这可能会降低细胞生产所需产品的能力。在这里,我们提出了一个与计算流体动力学相关的基于代理的数据驱动模型,最终允许预测大肠杆菌的额外 ATP 需求K12 W3110 暴露于真实的大型生物反应器条件。复杂模型描述了大约 600 个基因的转录上调和下调动态,从覆盖 28 小时的亚分钟范围开始。基于数据的方法是从全面的缩小实验中提取的。在 54 m 3搅拌生物反应器中模拟混合和传质条件,120,000大肠杆菌在不同葡萄糖可用性区域之间波动时跟踪细胞。结果表明,细胞 ATP 需求上升了增长解耦维持需求的 30% 至 45%,这可能会相应地限制 ATP 密集型产品形成的生产。此外,单个细胞转录模式的空间分析揭示了非常异质的基因扩增,在生物反应器的上部区域有 50%–80% 的信使 RNA 上调热点。这种现象反映了通过搅拌罐传播的细胞的延时调节反应。4.2 小时后,细胞适应环境变化,但仍需承受额外的 6% ATP 需求。


























 京公网安备 11010802027423号
京公网安备 11010802027423号