Journal of Proteomics ( IF 3.3 ) Pub Date : 2020-09-17 , DOI: 10.1016/j.jprot.2020.103988 Liam Cassidy 1 , Andreas O Helbig 1 , Philipp T Kaulich 1 , Kathrin Weidenbach 2 , Ruth A Schmitz 2 , Andreas Tholey 1
|
Short open reading frame-encoded peptides (SEP) represent a widely undiscovered part of the proteome. The detailed analysis of SEP has, despite inherent limitations such as incomplete sequence coverage, challenges encountered with protein inference, the identification of posttranslational modifications and the assignment of potential N- and C-terminal truncations, predominantly been assessed using bottom-up proteomic workflows. The use of top-down based proteomic workflows is capable of providing an unparalleled level of characterization information, which is of increased importance in the case of alternatively encoded protein products. However, top-down based analysis is not without its own limitations, for which efficient separation prior to MS analysis is a major issue.
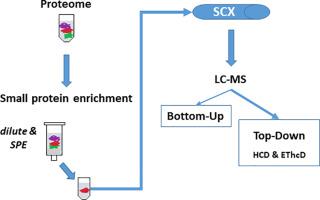
We established a sample preparation approach for the combined bottom-up and top-down proteomic analysis of SEP. Key improvements were made by the application of solid phase extraction (SPE), which supported enrichment of proteins below ca. 20 kDa, followed by 2D-LC-MS top-down analysis encompassing both HCD and EThcD ion activation. Bottom-up experiments were used to support and confirm top-down data interpretation. This strategy allowed for the top-down characterization of 36 proteoforms mapping to 12 SEP from the archaeon Methanosarcina mazei strain Gö1, with the concurrent detection and identification of several posttranslational modifications in SEP.
Biological significance
Small or short open reading frames (sORF) have been widely neglected in genome research in the past. With their increasing discovery, the question about the presence and molecular function of their translation products, the short open reading frame-encoded peptides (SEP), arises. As these small proteins are usually below the 10 kDa range, the number of peptides identifiable by bottom-up proteomics is limited which hampers both the identification and the recognition of potential posttranslational modifications. The presented top-down approach allowed for the detection of full length SEP, as well as of terminally truncated proteoforms, and further enabled the identification of disulfide bonds in these small proteins. This demonstrates, that this yet widely undiscovered part of the proteome undergoes the same modifications as classical proteins which is an essential step for future understanding of the biological functions of these molecules.
中文翻译:

多维分离方案可增强自上而下蛋白质组学中低分子量蛋白质组和短开放阅读框编码肽段的鉴定和分子表征。
短开放阅读框编码的肽(SEP)代表了蛋白质组中广泛未被发现的部分。尽管存在固有局限性,例如序列覆盖范围不完整,蛋白推断遇到的挑战,翻译后修饰的鉴定以及潜在的N和C端截短的分配等,但SEP的详细分析仍主要使用自下而上的蛋白质组学工作流程进行评估。基于自上而下的蛋白质组学工作流程的使用能够提供无与伦比的表征信息水平,这在交替编码的蛋白质产品中变得越来越重要。但是,基于自上而下的分析并非没有其自身的局限性,为此,MS分析之前的有效分离是一个主要问题。
我们为SEP的自下而上和自上而下的蛋白质组学分析建立了样品制备方法。固相萃取(SPE)的应用已取得了关键性的进步,固相萃取可支持蛋白质的富集。20 kDa,然后进行2D-LC-MS自上而下的分析,包括HCD和EThcD离子活化。自下而上的实验被用来支持和确认自上而下的数据解释。这种策略允许自上而下表征从古生甲烷八叠球菌菌株Gö1映射到12个SEP的36种蛋白形式,并同时检测和鉴定SEP中的几种翻译后修饰。
生物学意义
过去,小型或短开放阅读框(sORF)在基因组研究中被广泛忽略。随着发现的增加,出现了有关其翻译产物短开放阅读框编码肽(SEP)的存在和分子功能的问题。由于这些小蛋白质通常低于10 kDa,因此自下而上的蛋白质组学可识别的肽数量有限,这会阻碍鉴定和识别潜在的翻译后修饰。提出的自上而下的方法允许检测全长SEP以及末端截短的蛋白形式,并进一步使得能够鉴定这些小蛋白质中的二硫键。这表明


























 京公网安备 11010802027423号
京公网安备 11010802027423号