当前位置:
X-MOL 学术
›
Ecol. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Comparative assessment of range‐wide patterns of genetic diversity and structure with SNPs and microsatellites: A case study with Iberian amphibians
Ecology and Evolution ( IF 2.6 ) Pub Date : 2020-09-15 , DOI: 10.1002/ece3.6670 Miguel Camacho‐Sanchez 1 , Guillermo Velo‐Antón 1 , Jeffrey O. Hanson 1 , Ana Veríssimo 1 , Íñigo Martínez‐Solano 2 , Adam Marques 1 , Craig Moritz 3 , Sílvia B. Carvalho 1
Ecology and Evolution ( IF 2.6 ) Pub Date : 2020-09-15 , DOI: 10.1002/ece3.6670 Miguel Camacho‐Sanchez 1 , Guillermo Velo‐Antón 1 , Jeffrey O. Hanson 1 , Ana Veríssimo 1 , Íñigo Martínez‐Solano 2 , Adam Marques 1 , Craig Moritz 3 , Sílvia B. Carvalho 1
Affiliation
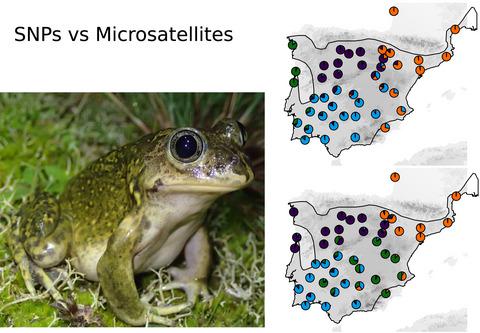
|
Reduced representation genome sequencing has popularized the application of single nucleotide polymorphisms (SNPs) to address evolutionary and conservation questions in nonmodel organisms. Patterns of genetic structure and diversity based on SNPs often diverge from those obtained with microsatellites to different degrees, but few studies have explicitly compared their performance under similar sampling regimes in a shared analytical framework. We compared range‐wide patterns of genetic structure and diversity in two amphibians endemic to the Iberian Peninsula: Hyla molleri and Pelobates cultripes, based on microsatellite (18 and 14 loci) and SNP (15,412 and 33,140 loci) datasets of comparable sample size and spatial extent. Model‐based clustering analyses with STRUCTURE revealed minor differences in genetic structure between marker types, but inconsistent values of the optimal number of populations (K) inferred. SNPs yielded more repeatable and less admixed ancestries with increasing K compared to microsatellites. Genetic diversity was weakly correlated between marker types, with SNPs providing a better representation of southern refugia and of gradients of genetic diversity congruent with the demographic history of both species. Our results suggest that the larger number of loci in a SNP dataset can provide more reliable inferences of patterns of genetic structure and diversity than a typical microsatellite dataset, at least at the spatial and temporal scales investigated.
中文翻译:

单核苷酸多态性和微卫星的遗传多样性和结构的全范围模式的比较评估:以伊比利亚两栖动物为例
减少代表性的基因组测序已普及了单核苷酸多态性(SNP)的应用,以解决非模型生物中的进化和保守问题。基于单核苷酸多态性的遗传结构和多样性模式通常与通过微卫星获得的模式有不同程度的差异,但是很少有研究在共享的分析框架中明确比较它们在相似采样模式下的性能。我们比较了伊比利亚半岛特有的两种两栖动物的遗传结构和多样性的全范围模式:Hyla molleri和Pelobates cultripes,基于可比较的样本大小和空间范围的微卫星(18和14个基因座)和SNP(15,412和33,140个基因座)数据集。使用STRUCTURE进行的基于模型的聚类分析显示,标记物类型之间的遗传结构存在细微差异,但推断出的最佳种群数(K)却不一致。与微卫星相比,随着K的增加,SNP产生更多的可重复性和更少的混合祖先。遗传多样性在标记物类型之间的相关性较弱,SNP可更好地代表南部避难所以及与这两个物种的人口历史一致的遗传多样性梯度。我们的结果表明,与典型的微卫星数据集相比,SNP数据集中更多的基因座可以提供更可靠的遗传结构和多样性模式推断,
更新日期:2020-10-12
中文翻译:

单核苷酸多态性和微卫星的遗传多样性和结构的全范围模式的比较评估:以伊比利亚两栖动物为例
减少代表性的基因组测序已普及了单核苷酸多态性(SNP)的应用,以解决非模型生物中的进化和保守问题。基于单核苷酸多态性的遗传结构和多样性模式通常与通过微卫星获得的模式有不同程度的差异,但是很少有研究在共享的分析框架中明确比较它们在相似采样模式下的性能。我们比较了伊比利亚半岛特有的两种两栖动物的遗传结构和多样性的全范围模式:Hyla molleri和Pelobates cultripes,基于可比较的样本大小和空间范围的微卫星(18和14个基因座)和SNP(15,412和33,140个基因座)数据集。使用STRUCTURE进行的基于模型的聚类分析显示,标记物类型之间的遗传结构存在细微差异,但推断出的最佳种群数(K)却不一致。与微卫星相比,随着K的增加,SNP产生更多的可重复性和更少的混合祖先。遗传多样性在标记物类型之间的相关性较弱,SNP可更好地代表南部避难所以及与这两个物种的人口历史一致的遗传多样性梯度。我们的结果表明,与典型的微卫星数据集相比,SNP数据集中更多的基因座可以提供更可靠的遗传结构和多样性模式推断,



























 京公网安备 11010802027423号
京公网安备 11010802027423号