当前位置:
X-MOL 学术
›
Immunology
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Elucidating host–microbe interactions in vivo by studying population dynamics using neutral genetic tags
Immunology ( IF 6.4 ) Pub Date : 2020-09-15 , DOI: 10.1111/imm.13266 Annika Hausmann 1 , Wolf-Dietrich Hardt 1
Immunology ( IF 6.4 ) Pub Date : 2020-09-15 , DOI: 10.1111/imm.13266 Annika Hausmann 1 , Wolf-Dietrich Hardt 1
Affiliation
|
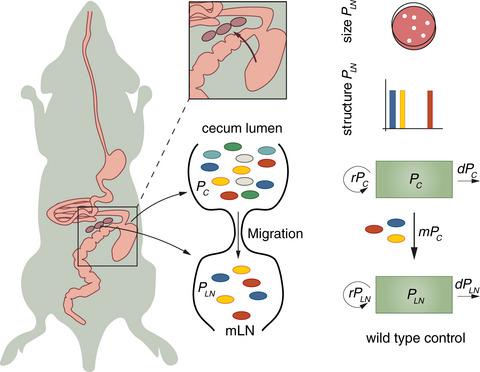
Host–microbe interactions are highly dynamic in space and time, in particular in the case of infections. Pathogen population sizes, microbial phenotypes and the nature of the host responses often change dramatically over time. These features pose particular challenges when deciphering the underlying mechanisms of these interactions experimentally, as traditional microbiological and immunological methods mostly provide snapshots of population sizes or sparse time series. Recent approaches – combining experiments using neutral genetic tags with stochastic population dynamic models – allow more precise quantification of biologically relevant parameters that govern the interaction between microbe and host cell populations. This is accomplished by exploiting the patterns of change of tag composition in the microbe or host cell population under study. These models can be used to predict the effects of immunodeficiencies or therapies (e.g. antibiotic treatment) on populations and thereby generate hypotheses and refine experimental designs. In this review, we present tools to study population dynamics in vivo using genetic tags, explain examples for their implementation and briefly discuss future applications.
中文翻译:

通过使用中性遗传标签研究种群动态来阐明体内宿主-微生物的相互作用
宿主-微生物相互作用在空间和时间上是高度动态的,特别是在感染的情况下。病原体种群大小、微生物表型和宿主反应的性质通常会随着时间发生显着变化。在通过实验破译这些相互作用的潜在机制时,这些特征构成了特别的挑战,因为传统的微生物学和免疫学方法主要提供种群规模或稀疏时间序列的快照。最近的方法——将使用中性遗传标签的实验与随机种群动态模型相结合——允许更精确地量化控制微生物和宿主细胞种群之间相互作用的生物学相关参数。这是通过利用所研究的微生物或宿主细胞群中标签组成的变化模式来实现的。这些模型可用于预测免疫缺陷或疗法(例如抗生素治疗)对人群的影响,从而产生假设并改进实验设计。在这篇综述中,我们提出了研究人口动态的工具在体内使用遗传标签,解释其实施的例子并简要讨论未来的应用。
更新日期:2020-09-15
中文翻译:

通过使用中性遗传标签研究种群动态来阐明体内宿主-微生物的相互作用
宿主-微生物相互作用在空间和时间上是高度动态的,特别是在感染的情况下。病原体种群大小、微生物表型和宿主反应的性质通常会随着时间发生显着变化。在通过实验破译这些相互作用的潜在机制时,这些特征构成了特别的挑战,因为传统的微生物学和免疫学方法主要提供种群规模或稀疏时间序列的快照。最近的方法——将使用中性遗传标签的实验与随机种群动态模型相结合——允许更精确地量化控制微生物和宿主细胞种群之间相互作用的生物学相关参数。这是通过利用所研究的微生物或宿主细胞群中标签组成的变化模式来实现的。这些模型可用于预测免疫缺陷或疗法(例如抗生素治疗)对人群的影响,从而产生假设并改进实验设计。在这篇综述中,我们提出了研究人口动态的工具在体内使用遗传标签,解释其实施的例子并简要讨论未来的应用。

























 京公网安备 11010802027423号
京公网安备 11010802027423号