Applied Soft Computing ( IF 8.7 ) Pub Date : 2020-09-12 , DOI: 10.1016/j.asoc.2020.106718 Saeed Pirmoradi , Mohammad Teshnehlab , Nosratollah Zarghami , Arash Sharifi
|
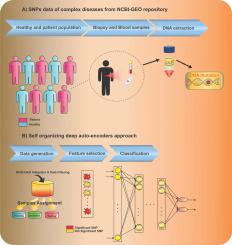
Recently, many Machine Learning algorithms have been utilized to identify significant Single Nucleotide Polymorphisms (SNPs) in various human diseases. However, some principal obstacles are challenging in the field of SNP detection and healthy-patient classification. The curse of dimensionality is the main challenge. On the other hand, the number of samples is decidedly smaller than the number of SNPs. In addition, the number of healthy and patient samples can be unequal. These challenges make the feature selection and classification very difficult. The main goal of the current study is the combination of the various algorithms to find out the most effective way of SNP data analysis. Therefore, an efficient method is proposed to identify significant SNPs and classify healthy and patient samples. In this regard, firstly, the Mean Encoding, as an intelligent method, is utilized to convert the nominal SNP data to numeric. Then a two-step filter method is used for feature selection, which removes the irrelevant and redundant features. Finally, the proposed deep auto-encoder is employed to classify so that it can construct its structure based on input data, automatically. To evaluate, we apply the proposed approach to five different SNP datasets, including thyroid cancer, mental retardation, breast cancer, colorectal cancer, and autism, which obtained from the Gene Expression Omnibus (GEO) dataset. The proposed method has succeeded in feature selection and classification so that it can classify healthy and patient samples based on selected features in thyroid cancer, mental retardation, breast cancer, colorectal cancer, and autism with 100%, 94.4%, 100%, 96%, and 99.1% accuracy, respectively. The results indicate that it has succeeded with high efficiency, compared with other published works.
中文翻译:

使用SNP基因组数据对复杂疾病进行分类的自组织深度自动编码器方法
最近,许多机器学习算法已被用于识别各种人类疾病中的重要单核苷酸多态性(SNP)。但是,在SNP检测和健康患者分类领域,一些主要障碍面临挑战。维数的诅咒是主要挑战。另一方面,样本数明显小于SNP数。此外,健康样本和患者样本的数量可能不相等。这些挑战使特征选择和分类非常困难。当前研究的主要目标是各种算法的组合,以找到最有效的SNP数据分析方法。因此,提出了一种有效的方法来鉴定重要的SNP并对健康和患者样品进行分类。在这方面,首先,均值编码 作为一种智能方法,可将名义SNP数据转换为数字。然后,采用两步过滤方法进行特征选择,以去除不相关和冗余的特征。最后,采用提出的深度自动编码器进行分类,以便它可以根据输入数据自动构造其结构。为了进行评估,我们将提议的方法应用于五个不同的SNP数据集,包括从基因表达综合(GEO)数据集获得的甲状腺癌,智力低下,乳腺癌,结肠直肠癌和自闭症。所提出的方法已经成功地进行了特征选择和分类,因此可以基于甲状腺癌,智力低下,乳腺癌,结肠直肠癌和自闭症中的选定特征对健康和患者样本进行分类,分别为100%,94.4%,100%,96% ,并且99.1%的准确性,分别。结果表明,与其他已发表的作品相比,它取得了成功。



























 京公网安备 11010802027423号
京公网安备 11010802027423号