当前位置:
X-MOL 学术
›
Hum. Mutat.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Cytogenetically visible inversions are formed by multiple molecular mechanisms.
Human Mutation ( IF 3.9 ) Pub Date : 2020-09-09 , DOI: 10.1002/humu.24106 Maria Pettersson 1, 2 , Christopher M Grochowski 3 , Josephine Wincent 1, 2 , Jesper Eisfeldt 1, 4 , Amy M Breman 5 , Sau W Cheung 3 , Ana C V Krepischi 6 , Carla Rosenberg 6 , James R Lupski 3, 7, 8 , Jesper Ottosson 9 , Lovisa Lovmar 9 , Jelena Gacic 10 , Elisabeth S Lundberg 1, 2 , Daniel Nilsson 1, 2, 4 , Claudia M B Carvalho 3, 11 , Anna Lindstrand 1, 2
Human Mutation ( IF 3.9 ) Pub Date : 2020-09-09 , DOI: 10.1002/humu.24106 Maria Pettersson 1, 2 , Christopher M Grochowski 3 , Josephine Wincent 1, 2 , Jesper Eisfeldt 1, 4 , Amy M Breman 5 , Sau W Cheung 3 , Ana C V Krepischi 6 , Carla Rosenberg 6 , James R Lupski 3, 7, 8 , Jesper Ottosson 9 , Lovisa Lovmar 9 , Jelena Gacic 10 , Elisabeth S Lundberg 1, 2 , Daniel Nilsson 1, 2, 4 , Claudia M B Carvalho 3, 11 , Anna Lindstrand 1, 2
Affiliation
|
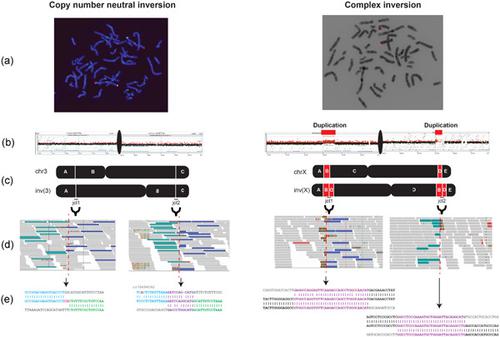
Cytogenetically detected inversions are generally assumed to be copy number and phenotypically neutral events. While nonallelic homologous recombination is thought to play a major role, recent data suggest the involvement of other molecular mechanisms in inversion formation. Using a combination of short‐read whole‐genome sequencing (WGS), 10X Genomics Chromium WGS, droplet digital polymerase chain reaction and array comparative genomic hybridization we investigated the genomic structure of 18 large unique cytogenetically detected chromosomal inversions and achieved nucleotide resolution of at least one chromosomal inversion junction for 13/18 (72%). Surprisingly, we observed that seemingly copy number neutral inversions can be accompanied by a copy‐number gain of up to 350 kb and local genomic complexities (3/18, 17%). In the resolved inversions, the mutational signatures are consistent with nonhomologous end‐joining (8/13, 62%) or microhomology‐mediated break‐induced replication (5/13, 38%). Our study indicates that short‐read 30x coverage WGS can detect a substantial fraction of chromosomal inversions. Moreover, replication‐based mechanisms are responsible for approximately 38% of those events leading to a significant proportion of inversions that are actually accompanied by additional copy‐number variation potentially contributing to the overall phenotypic presentation of those patients.
中文翻译:

细胞遗传学上可见的倒位是由多种分子机制形成的。
细胞遗传学检测到的倒位通常被认为是拷贝数和表型中性事件。虽然非等位基因同源重组被认为起主要作用,但最近的数据表明其他分子机制参与了倒位形成。使用短读长全基因组测序 (WGS)、10X Genomics Chromium WGS、液滴数字聚合酶链反应和阵列比较基因组杂交的组合,我们研究了 18 个大型独特的细胞遗传学检测染色体倒位的基因组结构,并实现了至少为13/18 (72%) 的一个染色体倒位连接。令人惊讶的是,我们观察到看似拷贝数中性倒置可以伴随高达 350 kb 的拷贝数增益和局部基因组复杂性 (3/18, 17%)。在已解决的反转中,突变特征与非同源末端连接 (8/13, 62%) 或微同源介导的断裂诱导复制 (5/13, 38%) 一致。我们的研究表明,短读长 30 倍覆盖 WGS 可以检测到很大一部分染色体倒位。此外,基于复制的机制导致大约 38% 的这些事件导致很大比例的倒置,这些倒置实际上伴随着额外的拷贝数变异,这些变异可能有助于这些患者的整体表型表现。
更新日期:2020-10-30
中文翻译:

细胞遗传学上可见的倒位是由多种分子机制形成的。
细胞遗传学检测到的倒位通常被认为是拷贝数和表型中性事件。虽然非等位基因同源重组被认为起主要作用,但最近的数据表明其他分子机制参与了倒位形成。使用短读长全基因组测序 (WGS)、10X Genomics Chromium WGS、液滴数字聚合酶链反应和阵列比较基因组杂交的组合,我们研究了 18 个大型独特的细胞遗传学检测染色体倒位的基因组结构,并实现了至少为13/18 (72%) 的一个染色体倒位连接。令人惊讶的是,我们观察到看似拷贝数中性倒置可以伴随高达 350 kb 的拷贝数增益和局部基因组复杂性 (3/18, 17%)。在已解决的反转中,突变特征与非同源末端连接 (8/13, 62%) 或微同源介导的断裂诱导复制 (5/13, 38%) 一致。我们的研究表明,短读长 30 倍覆盖 WGS 可以检测到很大一部分染色体倒位。此外,基于复制的机制导致大约 38% 的这些事件导致很大比例的倒置,这些倒置实际上伴随着额外的拷贝数变异,这些变异可能有助于这些患者的整体表型表现。


























 京公网安备 11010802027423号
京公网安备 11010802027423号