Water Research ( IF 12.8 ) Pub Date : 2020-09-09 , DOI: 10.1016/j.watres.2020.116397 Jiayu Zhang 1 , Wenhui Gan 2 , Renxin Zhao 1 , Ke Yu 3 , Huaxin Lei 1 , Ruiyang Li 1 , Xiaoyan Li 4 , Bing Li 2
|
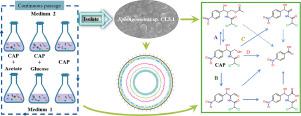
Figuring out the comprehensive metabolic mechanism of chloramphenicol (CAP) is critical to improving CAP removal in the bioremediation process. In this study, CAP biodegradation by six consortia and isolated Sphingomonas sp. CL5.1 were systematically investigated using the combination of high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry, second-generation, and third-generation sequencing technologies. The CAP-degrading capability of six consortia was enhanced while CAP mineralization rate declined after long-term enrichment. The microbial community structures of six consortia were all simplified with 69%−82% decline in species richness after continuous passages for one year. The core genera of consortia CL and CH included Sphingomonas, Cupriavidus, Burkholderia, Chryseobacterium, and Pigmentiphaga, which accounted for over 98% of the total population. Sphingomonas was discovered as a new CAP degrader that could subsist on CAP as the sole carbon, nitrogen, and energy sources. Sphingomonas sp. CL5.1 was able to completely remove 120 mg/L CAP within 48 hours with a mineralization rate of 50.4%. The presence of acetate or nitrite could inhibit CAP metabolization by strain CL5.1. Four CAP metabolic pathways were constructed, including modification of the C3 hydroxyl group of CAP via acetylation, oxidization, dehydration and the bond cleavage between C1 and C2. C3 hydroxyl group dehydration and C1-C2 bond-cleavage were first reported regarding to CAP biotransformation. Strain CL5.1 played a core role in the consortia and was responsible for C3 hydroxyl oxidation, C3 dehydration, and C1-C2 bond cleavage. Genomic information of strain CL5.1 revealed the further mineralization pathways of downstream product p-nitrobenzoic acid via ortho- and meta-cleavage.
中文翻译:

氯霉素的富集细菌菌群和分离的菌株鞘氨醇单胞菌菌种的生物降解。CL5.1:新型生物降解途径的重建。
弄清楚氯霉素(CAP)的全面代谢机制对于改善生物修复过程中CAP的去除至关重要。在这项研究中,CAP由六个财团和孤立的鞘氨醇单胞菌属生物降解。使用高效液相色谱-四极杆飞行时间质谱,第二代和第三代测序技术的组合,系统地研究了CL5.1。长期富集后,六个财团的CAP降解能力增强,而CAP矿化率下降。连续经过一年后,六个财团的微生物群落结构都得到了简化,物种丰富度下降了69%-82%。财团CL和CH的核心属鞘氨醇单胞菌,铜杯菌,伯克霍尔德氏菌,金黄色葡萄球菌和色素皮膜杆菌,占总人口的98%以上。鞘氨醇单胞菌被发现是一种新的CAP降解剂,可以作为唯一的碳,氮和能源而存在于CAP上。鞘氨醇单胞菌 CL5.1能够在48小时内完全去除120 mg / L CAP,矿化率为50.4%。乙酸盐或亚硝酸盐的存在可以抑制菌株CL5.1引起的CAP代谢。构建了四个CAP代谢途径,包括通过乙酰化,氧化,脱水以及C 1和C 2之间的键断裂来修饰CAP的C 3羟基。C 3关于CAP生物转化,首先报道了羟基脱水和C 1 -C 2键断裂。CL5.1菌株在该财团中起核心作用,并负责C3羟基氧化,C3脱水和C1-C2键断裂。菌株CL5.1的基因组信息揭示了通过邻位和间位裂解的下游产物对硝基苯甲酸的进一步矿化途径。

























 京公网安备 11010802027423号
京公网安备 11010802027423号