当前位置:
X-MOL 学术
›
Genes Brain Behav.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A long non‐coding RNA (Lrap) modulates brain gene expression and levels of alcohol consumption in rats
Genes, Brain and Behavior ( IF 2.5 ) Pub Date : 2020-09-06 , DOI: 10.1111/gbb.12698 Laura M Saba 1 , Paula L Hoffman 1, 2 , Gregg E Homanics 3 , Spencer Mahaffey 1 , Swapna Vidhur Daulatabad 4 , Sarath Chandra Janga 4, 5, 6 , Boris Tabakoff 1
Genes, Brain and Behavior ( IF 2.5 ) Pub Date : 2020-09-06 , DOI: 10.1111/gbb.12698 Laura M Saba 1 , Paula L Hoffman 1, 2 , Gregg E Homanics 3 , Spencer Mahaffey 1 , Swapna Vidhur Daulatabad 4 , Sarath Chandra Janga 4, 5, 6 , Boris Tabakoff 1
Affiliation
|
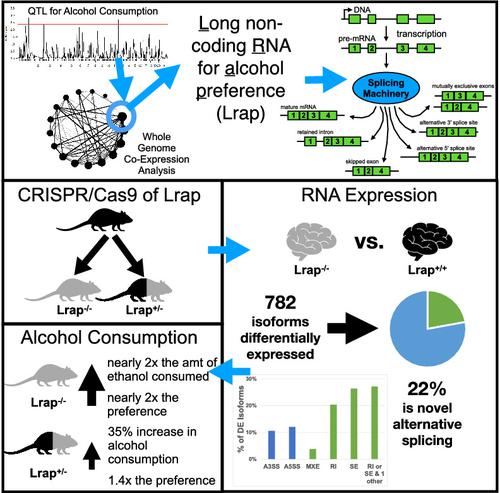
LncRNAs are important regulators of quantitative and qualitative features of the transcriptome. We have used QTL and other statistical analyses to identify a gene coexpression module associated with alcohol consumption. The “hub gene” of this module, Lrap (Long non‐coding RNA for alcohol preference), was an unannotated transcript resembling a lncRNA. We used partial correlation analyses to establish that Lrap is a major contributor to the integrity of the coexpression module. Using CRISPR/Cas9 technology, we disrupted an exon of Lrap in Wistar rats. Measures of alcohol consumption in wild type, heterozygous and knockout rats showed that disruption of Lrap produced increases in alcohol consumption/alcohol preference. The disruption of Lrap also produced changes in expression of over 700 other transcripts. Furthermore, it became apparent that Lrap may have a function in alternative splicing of the affected transcripts. The GO category of “Response to Ethanol” emerged as one of the top candidates in an enrichment analysis of the differentially expressed transcripts. We validate the role of Lrap as a mediator of alcohol consumption by rats, and also implicate Lrap as a modifier of the expression and splicing of a large number of brain transcripts. A defined subset of these transcripts significantly impacts alcohol consumption by rats (and possibly humans). Our work shows the pleiotropic nature of non‐coding elements of the genome, the power of network analysis in identifying the critical elements influencing phenotypes, and the fact that not all changes produced by genetic editing are critical for the concomitant changes in phenotype.
中文翻译:

长链非编码 RNA (Lrap) 调节大鼠大脑基因表达和饮酒水平
LncRNA 是转录组定量和定性特征的重要调节因子。我们使用 QTL 和其他统计分析来识别与饮酒相关的基因共表达模块。该模块的“中枢基因”Lrap(用于酒精偏好的长链非编码 RNA)是一个类似于 lncRNA 的未注释转录本。我们使用偏相关分析来确定 Lrap 是共表达模块完整性的主要贡献者。使用 CRISPR/Cas9 技术,我们破坏了 Wistar 大鼠中 Lrap 的一个外显子。野生型、杂合子和基因敲除大鼠的酒精消耗量度表明,Lrap 的破坏导致酒精消耗量/酒精偏好增加。Lrap 的破坏还导致 700 多个其他转录本的表达发生变化。此外,很明显,Lrap 可能在受影响转录本的可变剪接中发挥作用。“对乙醇的反应”的 GO 类别成为差异表达转录本富集分析中的最佳候选者之一。我们验证了 Lrap 作为大鼠饮酒中介的作用,并且还暗示 Lrap 作为大量大脑转录本表达和剪接的修饰剂。这些转录本的特定子集显着影响大鼠(可能还有人类)的饮酒量。我们的工作展示了基因组非编码元素的多效性、网络分析在识别影响表型的关键元素方面的能力,以及并非所有由基因编辑产生的变化都对伴随的表型变化至关重要的事实。
更新日期:2020-09-06
中文翻译:

长链非编码 RNA (Lrap) 调节大鼠大脑基因表达和饮酒水平
LncRNA 是转录组定量和定性特征的重要调节因子。我们使用 QTL 和其他统计分析来识别与饮酒相关的基因共表达模块。该模块的“中枢基因”Lrap(用于酒精偏好的长链非编码 RNA)是一个类似于 lncRNA 的未注释转录本。我们使用偏相关分析来确定 Lrap 是共表达模块完整性的主要贡献者。使用 CRISPR/Cas9 技术,我们破坏了 Wistar 大鼠中 Lrap 的一个外显子。野生型、杂合子和基因敲除大鼠的酒精消耗量度表明,Lrap 的破坏导致酒精消耗量/酒精偏好增加。Lrap 的破坏还导致 700 多个其他转录本的表达发生变化。此外,很明显,Lrap 可能在受影响转录本的可变剪接中发挥作用。“对乙醇的反应”的 GO 类别成为差异表达转录本富集分析中的最佳候选者之一。我们验证了 Lrap 作为大鼠饮酒中介的作用,并且还暗示 Lrap 作为大量大脑转录本表达和剪接的修饰剂。这些转录本的特定子集显着影响大鼠(可能还有人类)的饮酒量。我们的工作展示了基因组非编码元素的多效性、网络分析在识别影响表型的关键元素方面的能力,以及并非所有由基因编辑产生的变化都对伴随的表型变化至关重要的事实。


























 京公网安备 11010802027423号
京公网安备 11010802027423号