Journal of Structural Biology ( IF 3 ) Pub Date : 2020-09-05 , DOI: 10.1016/j.jsb.2020.107608 Lisanna Paladin 1 , Marco Necci 1 , Damiano Piovesan 1 , Pablo Mier 2 , Miguel A Andrade-Navarro 2 , Silvio C E Tosatto 1
|
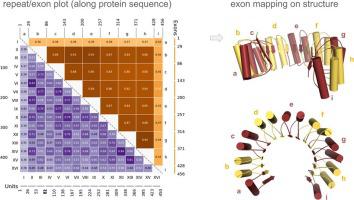
Tandem Repeat Proteins (TRPs) are ubiquitous in cells and are enriched in eukaryotes. They contributed to the evolution of organism complexity, specializing for functions that require quick adaptability such as immunity-related functions. To investigate the hypothesis of repeat protein evolution through exon duplication and rearrangement, we designed a tool to analyze the relationships between exon/intron patterns and structural symmetries. The tool allows comparison of the structure fragments as defined by exon/intron boundaries from Ensembl against the structural element repetitions from RepeatsDB. The all-against-all pairwise structural alignment between fragments and comparison of the two definitions (structural units and exons) are visualized in a single matrix, the “repeat/exon plot”. An analysis of different repeat protein families, including the solenoids Leucine-Rich, Ankyrin, Pumilio, HEAT repeats and the β propellers Kelch-like, WD40 and RCC1, shows different behaviors, illustrated here through examples. For each example, the analysis of the exon mapping in homologous proteins supports the conservation of their exon patterns. We propose that when a clear-cut relationship between exon and structural boundaries can be identified, it is possible to infer a specific “evolutionary pattern” which may improve TRPs detection and classification.
中文翻译:

一种通过外显子复制研究结构串联重复蛋白家族进化的新方法。
串联重复蛋白 (TRP) 在细胞中无处不在,并在真核生物中富集。它们有助于生物体复杂性的进化,专门用于需要快速适应性的功能,例如免疫相关功能。为了通过外显子复制和重排研究重复蛋白质进化的假设,我们设计了一个工具来分析外显子/内含子模式和结构对称性之间的关系。该工具允许将来自 Ensembl 的外显子/内含子边界定义的结构片段与来自 RepeatsDB 的结构元素重复进行比较。片段之间完全相反的成对结构对齐和两个定义(结构单位和外显子)的比较在单个矩阵中可视化,即“重复/外显子图”。不同重复蛋白家族的分析,包括富含亮氨酸的螺线管、Ankyrin、Pumilio、HEAT 重复和 β 螺旋桨 Kelch-like、WD40 和 RCC1,显示不同的行为,此处通过示例进行说明。对于每个例子,同源蛋白质的外显子映射分析支持其外显子模式的保守性。我们建议,当可以识别外显子和结构边界之间的明确关系时,就有可能推断出一种特定的“进化模式”,这可能会改善 TRP 的检测和分类。


























 京公网安备 11010802027423号
京公网安备 11010802027423号