当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Three-Dimensional Mass Spectrometric Imaging of Biological Structures Using a Vacuum-Compatible Microfluidic Device.
Analytical Chemistry ( IF 7.4 ) Pub Date : 2020-09-01 , DOI: 10.1021/acs.analchem.0c02204 Wenxiao Guo 1, 2 , Michal Kanski 3 , Wen Liu 1 , Mikołaj Gołuński 3 , Yadong Zhou 1 , Yining Wang 4 , Cuixia Cheng 1 , Yingge Du 4 , Zbigniew Postawa 3 , Wei David Wei 2 , Zihua Zhu 1
Analytical Chemistry ( IF 7.4 ) Pub Date : 2020-09-01 , DOI: 10.1021/acs.analchem.0c02204 Wenxiao Guo 1, 2 , Michal Kanski 3 , Wen Liu 1 , Mikołaj Gołuński 3 , Yadong Zhou 1 , Yining Wang 4 , Cuixia Cheng 1 , Yingge Du 4 , Zbigniew Postawa 3 , Wei David Wei 2 , Zihua Zhu 1
Affiliation
|
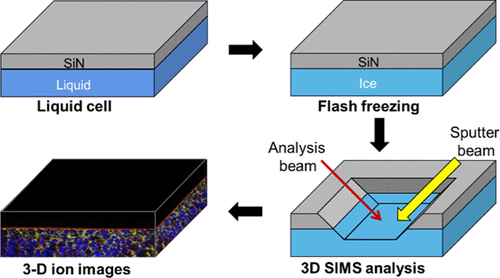
Three-dimensional (3D) molecular imaging of biological structures is important for a wide range of research. In recent decades, secondary-ion mass spectrometry (SIMS) has been recognized as a powerful technique for both two-dimensional and 3D molecular imaging. Sample fixations (e.g., chemical fixation and cryogenic fixation methods) are necessary to adapt biological samples to the vacuum condition in the SIMS chamber, which has been demonstrated to be nontrivial and less controllable, thus limiting the wider application of SIMS on 3D molecular analysis of biological samples. Our group recently developed in situ liquid SIMS that offers great opportunities for the molecular study of various liquids and liquid interfaces. In this work, we demonstrate that a further development of the vacuum-compatible microfluidic device used in in situ liquid SIMS provides a convenient freeze-fixation of biological samples and leads to more controllable and convenient 3D molecular imaging. The special design of this new vacuum-compatible liquid chamber allows an easy determination of sputter rates of ice, which is critical for calibrating the depth scale of frozen biological samples. Sputter yield of a 20 keV Ar1800+ ion on ice has been determined as 1500 (±8%) water molecules per Ar1800+ ion, consistent with our results from molecular dynamics simulations. Moreover, using the information of ice sputter yield, we successfully conduct 3D molecular imaging of frozen homogenized milk and observe network structures of interesting organic and inorganic species. Taken together, our results will significantly benefit various research fields relying on 3D molecular imaging of biological structures.
中文翻译:

使用真空兼容的微流控设备对生物结构进行三维质谱成像。
生物结构的三维(3D)分子成像对于广泛的研究非常重要。在最近的几十年中,二次离子质谱(SIMS)被公认为是二维和3D分子成像的强大技术。样品固定(例如化学固定和低温固定方法)对于使生物样品适应SIMS腔室中的真空条件是必需的,这已证明是不平凡且不可控的,因此限制了SIMS在3D分子分析中的广泛应用生物样品。我们小组最近在原地发展液体SIMS为各种液体和液体界面的分子研究提供了巨大的机会。在这项工作中,我们证明了原位液体SIMS中使用的与真空兼容的微流体装置的进一步发展为生物样品的冷冻固定提供了便利,并导致了可控和便利的3D分子成像。这种新型的与真空兼容的液体腔室的特殊设计可以轻松确定冰的溅射速率,这对于校准冷冻生物样品的深度刻度至关重要。已确定在冰上20 keV Ar 1800 +离子的溅射产率为每Ar 1800 + 1500(±8%)个水分子离子,与我们从分子动力学模拟得出的结果一致。此外,利用冰溅射产量的信息,我们成功地进行了冷冻均质牛奶的3D分子成像,并观察了有趣的有机和无机物种的网络结构。综上所述,我们的结果将极大地有益于依赖于生物结构的3D分子成像的各个研究领域。
更新日期:2020-10-21
中文翻译:

使用真空兼容的微流控设备对生物结构进行三维质谱成像。
生物结构的三维(3D)分子成像对于广泛的研究非常重要。在最近的几十年中,二次离子质谱(SIMS)被公认为是二维和3D分子成像的强大技术。样品固定(例如化学固定和低温固定方法)对于使生物样品适应SIMS腔室中的真空条件是必需的,这已证明是不平凡且不可控的,因此限制了SIMS在3D分子分析中的广泛应用生物样品。我们小组最近在原地发展液体SIMS为各种液体和液体界面的分子研究提供了巨大的机会。在这项工作中,我们证明了原位液体SIMS中使用的与真空兼容的微流体装置的进一步发展为生物样品的冷冻固定提供了便利,并导致了可控和便利的3D分子成像。这种新型的与真空兼容的液体腔室的特殊设计可以轻松确定冰的溅射速率,这对于校准冷冻生物样品的深度刻度至关重要。已确定在冰上20 keV Ar 1800 +离子的溅射产率为每Ar 1800 + 1500(±8%)个水分子离子,与我们从分子动力学模拟得出的结果一致。此外,利用冰溅射产量的信息,我们成功地进行了冷冻均质牛奶的3D分子成像,并观察了有趣的有机和无机物种的网络结构。综上所述,我们的结果将极大地有益于依赖于生物结构的3D分子成像的各个研究领域。



























 京公网安备 11010802027423号
京公网安备 11010802027423号