Journal of Molecular Graphics and Modelling ( IF 2.9 ) Pub Date : 2020-09-02 , DOI: 10.1016/j.jmgm.2020.107730 Ikechukwu Achilonu 1 , Emmanuel Amarachi Iwuchukwu 1 , Okechinyere Juliet Achilonu 2 , Manuel Antonio Fernandes 3 , Yasien Sayed 1
|
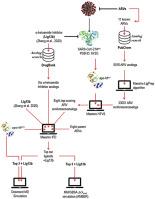
The SARS-CoV-2 main protease (Mpro) is an attractive target towards discovery of drugs to treat COVID-19 because of its key role in virus replication. The atomic structure of Mpro in complex with an α-ketoamide inhibitor (Lig13b) is available (PDB ID:6Y2G). Using 6Y2G and the prior knowledge that protease inhibitors could eradicate COVID-19, we designed a computational study aimed at identifying FDA-approved drugs that could interact with Mpro. We searched the DrugBank and PubChem for analogs and built a virtual library containing ∼33,000 conformers. Using high-throughput virtual screening and ligand docking, we identified Isavuconazonium, a ketoamide inhibitor (α-KI) and Pentagastrin as the top three molecules (Lig13b as the benchmark) based on docking energy. The ΔGbind of Lig13b, Isavuconazonium, α-KI, Pentagastrin was −28.1, −45.7, −44.7, −34.8 kcal/mol, respectively. Molecular dynamics simulation revealed that these ligands are stable within the Mpro active site. Binding of these ligands is driven by a variety of non-bonded interaction, including polar bonds, H-bonds, van der Waals and salt bridges. The overall conformational dynamics of the complexed-Mpro was slightly altered relative to apo-Mpro. This study demonstrates that three distinct classes molecules, Isavuconazonium (triazole), α-KI (ketoamide) and Pentagastrin (peptide) could serve as potential drugs to treat patients with COVID-19.
中文翻译:

使用FDA批准的Isavuconazonium,P2-P3α-酮酰胺衍生物和Pentagastrin靶向SARS-CoV-2主蛋白酶:一种计算机内药物发现方法。
SARS-CoV-2主蛋白酶(M pro)由于发现其在病毒复制中的关键作用而成为发现治疗COVID-19的药物的诱人靶标。M pro与α-酮酰胺抑制剂(Lig13b)形成复合物的原子结构可用(PDB ID:6Y2G)。利用6Y2G和蛋白酶抑制剂可以消除COVID-19的先验知识,我们设计了一项计算研究,旨在鉴定可与M pro相互作用的FDA批准药物。我们在DrugBank和PubChem中搜索了类似物,并建立了一个包含约33,000个构象体的虚拟文库。使用高通量虚拟筛选和配体对接,基于对接能,我们确定了Isavuconazonium,一种酮酰胺抑制剂(α-KI)和Pentagastrin作为排名前三的分子(以Lig13b为基准)。该Δ ģ绑定Lig13b,Isavuconazonium,α-KI的,五肽胃泌素是-28.1,-45.7,-44.7,-34.8千卡/摩尔,分别。分子动力学模拟表明,这些配体在M pro活性位点内是稳定的。这些配体的结合受多种非键相互作用的驱动,包括极性键,氢键,范德华力和盐桥。Complexed-M pro的整体构象动力学相对于apo-M pro略有改变。这项研究表明,三个不同类别的分子,Isavuconazonium(三唑),α-KI(酮酰胺)和Pentagastrin(肽)可以用作治疗COVID-19患者的潜在药物。



























 京公网安备 11010802027423号
京公网安备 11010802027423号