当前位置:
X-MOL 学术
›
Cryst. Growth Des.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Machine-Learning-Guided Cocrystal Prediction Based on Large Data Base
Crystal Growth & Design ( IF 3.8 ) Pub Date : 2020-08-31 , DOI: 10.1021/acs.cgd.0c00767 Dingyan Wang 1, 2 , Zeen Yang 2, 3 , Bingqing Zhu 3 , Xuefeng Mei 2, 3 , Xiaomin Luo 1, 2
Crystal Growth & Design ( IF 3.8 ) Pub Date : 2020-08-31 , DOI: 10.1021/acs.cgd.0c00767 Dingyan Wang 1, 2 , Zeen Yang 2, 3 , Bingqing Zhu 3 , Xuefeng Mei 2, 3 , Xiaomin Luo 1, 2
Affiliation
|
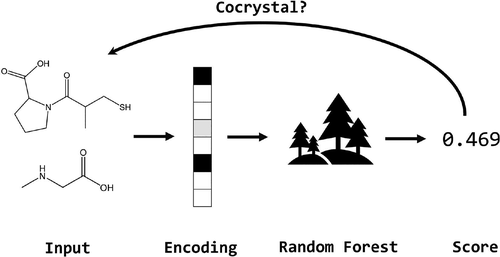
A machine-learning model trained on the whole Cambridge Structural Database was developed to assist high-throughput cocrystal screening. With only 2D structures taken as inputs, the probability of cocrystal formation is returned for two given molecules. All of the cocrystal records in the CSD were used as positive samples, while negative samples were constructed by randomly combining different molecules into chemical pairs. Our model showed a prediction ability comparable with that of a widely used ab initio method in a head-to-head comparison test. Both experimental and virtual cocrystal screening against captopril were conducted at the same time to further validate the model. Two cocrystals of captopril with l-proline and sarcosine were obtained and characterized by PXRD, DSC, and FT-IR. These two coformers were also successfully predicted by our model. These results suggest that the tool we developed can be used to effectively guide coformer selection in the discovery of new cocrystals.
中文翻译:

基于大数据库的机器学习指导共晶预测
开发了在整个剑桥结构数据库上训练的机器学习模型,以辅助高通量共晶筛选。仅将2D结构作为输入,对于两个给定的分子,共晶形成的可能性就会返回。CSD中所有共晶记录均用作阳性样品,而阴性样品则通过将不同分子随机组合成化学对而构建。我们的模型在头对头比较测试中显示了与广泛使用的从头算方法相当的预测能力。同时对卡托普利进行了实验和虚拟共晶筛选,以进一步验证模型。卡托普利与l的两个共结晶获得了脯氨酸和肌氨酸,并通过PXRD,DSC和FT-IR对其进行了表征。我们的模型也成功地预测了这两个同伴。这些结果表明,我们开发的工具可用于有效地指导新共晶体发现中的共形成器选择。
更新日期:2020-10-07
中文翻译:

基于大数据库的机器学习指导共晶预测
开发了在整个剑桥结构数据库上训练的机器学习模型,以辅助高通量共晶筛选。仅将2D结构作为输入,对于两个给定的分子,共晶形成的可能性就会返回。CSD中所有共晶记录均用作阳性样品,而阴性样品则通过将不同分子随机组合成化学对而构建。我们的模型在头对头比较测试中显示了与广泛使用的从头算方法相当的预测能力。同时对卡托普利进行了实验和虚拟共晶筛选,以进一步验证模型。卡托普利与l的两个共结晶获得了脯氨酸和肌氨酸,并通过PXRD,DSC和FT-IR对其进行了表征。我们的模型也成功地预测了这两个同伴。这些结果表明,我们开发的工具可用于有效地指导新共晶体发现中的共形成器选择。



























 京公网安备 11010802027423号
京公网安备 11010802027423号