Bioorganic Chemistry ( IF 5.1 ) Pub Date : 2020-08-28 , DOI: 10.1016/j.bioorg.2020.104244 Priyanka Sharma 1 , Manoj Kumar 1 , Sushila Dahiya 2 , Seema Sood 2 , Bimal Kumar Das 2 , Punit Kaur 1 , Arti Kapil 2
|
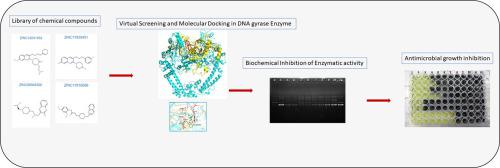
The emerged resistance in Typhoidal Salmonella has limited the treatment options for typhoid fever. In this scenario, there is a need to find alternate treatment modalities against this pathogen. Amongst the therapeutic agents currently being used to treat enteric fever, quinolones have enjoyed considerable success since past three decades. These drugs act upon DNA gyrase and the acquired resistance is due to mutations at Ser83 and Asp87 of gyrase A subunit. In the present study DNA gyrase enzyme was targeted to seek out potential new inhibitors which are not affected by these mutations. Molecular modelling and docking studies were performed in Schrödinger’s molecular modelling software. Homology model of DNA gyrase-DNA complex was built using templates 1AB4 and 3LTN. Molecular dynamic simulations were performed in SPC solvent for 100 ns. Total 17,900,742 drug like molecules were downloaded from ZINC library of chemical compounds. The Glide XP score of the compounds ranged from −5.285 to −13.692. All the ligands bound at the four base pair staggered nick in the DNA binding groove of DNA gyrase enzyme with their aromatic rings intercalating between the bases of two successive nucleotides stabilized by π - π stacking interactions. The binding pocket of DNA gyrase B comprising conserved residues Lys 447, Gly 448, Lys 449, Ile 450, Leu 451, Gln 465 and Val 467 interacts with the ligand molecules through van der Waals interactions. The MIC (minimum inhibitory concentration), MBC (minimum bactericidal concentration) and IC50 of the tested compounds ranged from 500 to 125 mg/L, 750 to 500 mg/L and 100 to 12.5 mg/L, respectively. The selected hits bind to quinolone binding pocket, but their mode of binding and conformation is different to fluoroquinolones, and hence, their binding is not affected by mutations at Ser83 or Asp87 positions. These lead compounds can be further explored as a scaffold to design inhibitors against DNA gyrase to bypass quinolone resistance.
中文翻译:

肠炎沙门氏菌伤寒沙门氏菌DNA促旋酶抑制剂的基于结构的药物发现和体外活性测试。
伤寒沙门氏菌中出现的耐药性限制了伤寒的治疗选择。在这种情况下,需要找到针对这种病原体的替代治疗方式。在目前用于治疗肠热的治疗剂中,喹诺酮类药物自过去的三十年以来已获得了相当大的成功。这些药物作用于DNA促旋酶,而获得的抗药性是由于促旋酶A亚基的Ser83和Asp87发生突变。在本研究中,以DNA促旋酶为目标,以寻找不受这些突变影响的潜在新抑制剂。分子建模和对接研究在Schrödinger的分子建模软件中进行。使用模板1AB4和3LTN构建DNA促旋酶-DNA复合物的同源性模型。在SPC溶剂中进行分子动力学模拟100 ns。总计17,900,从ZINC化合物库中下载了742种药物样分子。化合物的Glide XP得分在-5.285至-13.692之间。所有结合在四个碱基对上的配体在DNA促旋酶的DNA结合槽中交错排列,形成缺口,其芳香环插入通过π-π堆积相互作用稳定的两个连续核苷酸的碱基之间。包含保守残基Lys 447,Gly 448,Lys 449,Ile 450,Leu 451,Gln 465和Val 467的DNA促旋酶B的结合口袋通过范德华相互作用与配体分子相互作用。MIC(最小抑菌浓度),MBC(最小杀菌浓度)和IC 所有结合在四个碱基对上的配体在DNA促旋酶的DNA结合槽中交错排列,形成缺口,其芳香环插入通过π-π堆积相互作用稳定的两个连续核苷酸的碱基之间。包含保守残基Lys 447,Gly 448,Lys 449,Ile 450,Leu 451,Gln 465和Val 467的DNA促旋酶B的结合口袋通过范德华相互作用与配体分子相互作用。MIC(最小抑菌浓度),MBC(最小杀菌浓度)和IC 所有结合在四个碱基对上的配体在DNA促旋酶的DNA结合槽中交错排列,形成缺口,其芳香环插入通过π-π堆积相互作用稳定的两个连续核苷酸的碱基之间。包含保守残基Lys 447,Gly 448,Lys 449,Ile 450,Leu 451,Gln 465和Val 467的DNA促旋酶B的结合口袋通过范德华相互作用与配体分子相互作用。MIC(最小抑菌浓度),MBC(最小杀菌浓度)和IC Gln 465和Val 467通过范德华相互作用与配体分子相互作用。MIC(最小抑菌浓度),MBC(最小杀菌浓度)和IC Gln 465和Val 467通过范德华相互作用与配体分子相互作用。MIC(最小抑菌浓度),MBC(最小杀菌浓度)和IC50种受试化合物的浓度分别为500至125 mg / L,750至500 mg / L和100至12.5 mg / L。选定的命中结合到喹诺酮结合口袋,但它们的结合和构象模式不同于氟喹诺酮,因此,它们的结合不受Ser83或Asp87位置突变的影响。这些前导化合物可以作为支架进一步探索,以设计针对DNA促旋酶的抑制剂来绕过喹诺酮耐药性。



























 京公网安备 11010802027423号
京公网安备 11010802027423号