当前位置:
X-MOL 学术
›
Int. Biodeterior. Biodegrad.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Metagenomic characterization of microbial communities on plasticized fabric materials exposed to harsh tropical environments
International Biodeterioration & Biodegradation ( IF 4.8 ) Pub Date : 2020-10-01 , DOI: 10.1016/j.ibiod.2020.105061 Osman Radwan , Jason S. Lee , Robert Stote , Kevin Kuehn , Oscar N. Ruiz
International Biodeterioration & Biodegradation ( IF 4.8 ) Pub Date : 2020-10-01 , DOI: 10.1016/j.ibiod.2020.105061 Osman Radwan , Jason S. Lee , Robert Stote , Kevin Kuehn , Oscar N. Ruiz
|
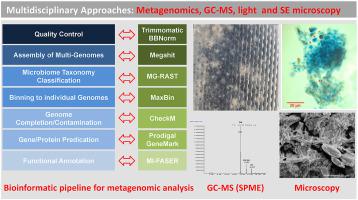
Abstract Biodeterioration of plasticized fabrics is a serious problem leading to degradation of materials used in military and civilian applications. This study aimed to characterize the composition of the microbial communities present on six plasticized fabrics exposed to a harsh tropical environment and explore their role in biodeterioration. Metagenomics, bioinformatics, light and scanning electron microscopy (SEM), and solid-phase microextraction GC-MS were used to characterize the fabric-associated microbial communities and plasticizer degradation. SEM analysis showed multi-layered biofilms containing bacteria and a high abundance of fungal structures and yeast cells. Shotgun metagenomics with a multifaceted bioinformatic pipeline generated 3,314,688 contigs and 120 microbial genomes. The microbial genomes were classified into three domains with the majority belonging to fungi followed by bacteria, and archaea. Functional gene annotation revealed that the fabric-associated microbiomes harbor important pathways for energy generation, stress tolerance, and compounds degradation including esterases, lipases and monooxygenases, suggesting a role of these microorganisms in degradation. The results showed that black yeasts were the keystone species of microbiomes affecting the fabrics. Information from this study will help to understand the effect of different microbial communities and species on the biodeterioration of fabrics and may help support the development of new antimicrobial materials.
中文翻译:

暴露于恶劣热带环境的增塑织物材料上微生物群落的宏基因组特征
摘要 增塑织物的生物降解是导致军事和民用应用材料降解的严重问题。本研究旨在表征暴露于恶劣热带环境的六种增塑织物上微生物群落的组成,并探索它们在生物退化中的作用。宏基因组学、生物信息学、光学和扫描电子显微镜 (SEM) 以及固相微萃取 GC-MS 用于表征与织物相关的微生物群落和增塑剂降解。SEM 分析显示多层生物膜含有细菌和大量真菌结构和酵母细胞。具有多方面生物信息学管道的 Shotgun 宏基因组学生成了 3,314,688 个重叠群和 120 个微生物基因组。微生物基因组分为三个域,其中大部分属于真菌,其次是细菌和古细菌。功能基因注释显示,与织物相关的微生物组具有重要的能量产生、胁迫耐受性和化合物降解途径,包括酯酶、脂肪酶和单加氧酶,表明这些微生物在降解中发挥作用。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。功能基因注释显示,与织物相关的微生物组具有重要的能量产生、胁迫耐受性和化合物降解途径,包括酯酶、脂肪酶和单加氧酶,表明这些微生物在降解中发挥作用。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。功能基因注释显示,与织物相关的微生物组具有重要的能量产生、胁迫耐受性和化合物降解途径,包括酯酶、脂肪酶和单加氧酶,表明这些微生物在降解中发挥作用。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。
更新日期:2020-10-01
中文翻译:

暴露于恶劣热带环境的增塑织物材料上微生物群落的宏基因组特征
摘要 增塑织物的生物降解是导致军事和民用应用材料降解的严重问题。本研究旨在表征暴露于恶劣热带环境的六种增塑织物上微生物群落的组成,并探索它们在生物退化中的作用。宏基因组学、生物信息学、光学和扫描电子显微镜 (SEM) 以及固相微萃取 GC-MS 用于表征与织物相关的微生物群落和增塑剂降解。SEM 分析显示多层生物膜含有细菌和大量真菌结构和酵母细胞。具有多方面生物信息学管道的 Shotgun 宏基因组学生成了 3,314,688 个重叠群和 120 个微生物基因组。微生物基因组分为三个域,其中大部分属于真菌,其次是细菌和古细菌。功能基因注释显示,与织物相关的微生物组具有重要的能量产生、胁迫耐受性和化合物降解途径,包括酯酶、脂肪酶和单加氧酶,表明这些微生物在降解中发挥作用。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。功能基因注释显示,与织物相关的微生物组具有重要的能量产生、胁迫耐受性和化合物降解途径,包括酯酶、脂肪酶和单加氧酶,表明这些微生物在降解中发挥作用。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。功能基因注释显示,与织物相关的微生物组具有重要的能量产生、胁迫耐受性和化合物降解途径,包括酯酶、脂肪酶和单加氧酶,表明这些微生物在降解中发挥作用。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。结果表明,黑酵母是影响织物的微生物群落的关键物种。这项研究的信息将有助于了解不同微生物群落和物种对织物生物降解的影响,并可能有助于支持新抗菌材料的开发。


























 京公网安备 11010802027423号
京公网安备 11010802027423号