当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Quantifying the Relationship between Conformational Dynamics and Enzymatic Activity in Ribonuclease HI Homologues.
Biochemistry ( IF 2.9 ) Pub Date : 2020-08-19 , DOI: 10.1021/acs.biochem.0c00500 James A Martin 1 , Paul Robustelli 2 , Arthur G Palmer 3
Biochemistry ( IF 2.9 ) Pub Date : 2020-08-19 , DOI: 10.1021/acs.biochem.0c00500 James A Martin 1 , Paul Robustelli 2 , Arthur G Palmer 3
Affiliation
|
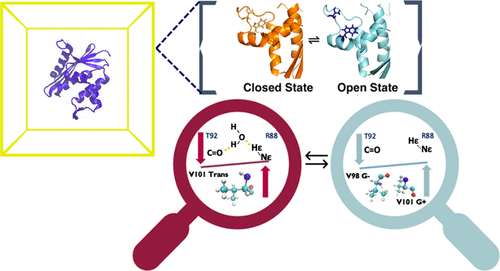
Ribonuclease HI (RNHI), a ubiquitous, non-sequence-specific endonuclease, cleaves the RNA strand in RNA/DNA hybrids. RNHI functions in replication and genome maintenance, and retroviral reverse transcriptases contain an essential ribonuclease H domain. Nuclear magnetic resonance (NMR) spectroscopy combined with molecular dynamics (MD) simulations suggests a model in which the extended handle region domain of Escherichia coli RNHI populates (substrate-binding-competent) “open” and (substrate-binding-incompetent) “closed” states, while the thermophilic Thermus thermophilus RNHI mainly populates the closed state at 300 K [Stafford, K. A., Robustelli, P., and Palmer, A. G., III (2013) PLoS Comput. Biol.9, 1–10]. In addition, an in silico-designed mutant E. coli Val98Ala RNHI was predicted to populate primarily the closed state. The work presented here validates this model and confirms the predicted properties of the designed mutant. MD simulations suggest that the conformational preferences of the handle region correlate with the conformations of Trp85, Thr92, and Val101. NMR residual dipolar coupling constants, three-bond scalar coupling constants, and chemical shifts experimentally define the conformational states of these residues and hence of the handle domain. These NMR parameters correlate with the Michaelis constants for RNHI homologues, confirming the important role of the handle region in the modulation of substrate recognition and illustrating the power of NMR spectroscopy in dissecting the conformational preferences underlying enzyme function.
中文翻译:

量化核糖核酸酶 HI 同源物中构象动力学与酶活性之间的关系。
核糖核酸酶 HI (RNHI) 是一种普遍存在的非序列特异性核酸内切酶,可切割 RNA/DNA 杂交体中的 RNA 链。RNHI 在复制和基因组维护中发挥作用,逆转录病毒逆转录酶包含一个必不可少的核糖核酸酶 H 结构域。核磁共振 (NMR) 光谱结合分子动力学 (MD) 模拟提出了一个模型,其中大肠杆菌RNHI的扩展手柄区域域填充(底物结合能力)“开放”和(底物结合不能力)“封闭” ” 状态,而嗜热的嗜热栖热菌RNHI 主要在 300 K 时处于封闭状态 [Stafford, KA, Robustelli, P., and Palmer, AG, III (2013) PLoS Comput. 生物。9 , 1–10]。此外,一个在硅-设计的突变大肠杆菌Val98Ala RNHI 预计主要填充封闭状态。此处介绍的工作验证了该模型并确认了设计突变体的预测特性。MD 模拟表明手柄区域的构象偏好与 Trp85、Thr92 和 Val101 的构象相关。NMR 残余偶极耦合常数、三键标量耦合常数和化学位移通过实验定义了这些残基的构象状态,从而定义了手柄域的构象状态。这些 NMR 参数与 RNHI 同系物的 Michaelis 常数相关,证实了手柄区域在调节底物识别中的重要作用,并说明了 NMR 光谱在剖析酶功能的构象偏好方面的能力。
更新日期:2020-09-08
中文翻译:

量化核糖核酸酶 HI 同源物中构象动力学与酶活性之间的关系。
核糖核酸酶 HI (RNHI) 是一种普遍存在的非序列特异性核酸内切酶,可切割 RNA/DNA 杂交体中的 RNA 链。RNHI 在复制和基因组维护中发挥作用,逆转录病毒逆转录酶包含一个必不可少的核糖核酸酶 H 结构域。核磁共振 (NMR) 光谱结合分子动力学 (MD) 模拟提出了一个模型,其中大肠杆菌RNHI的扩展手柄区域域填充(底物结合能力)“开放”和(底物结合不能力)“封闭” ” 状态,而嗜热的嗜热栖热菌RNHI 主要在 300 K 时处于封闭状态 [Stafford, KA, Robustelli, P., and Palmer, AG, III (2013) PLoS Comput. 生物。9 , 1–10]。此外,一个在硅-设计的突变大肠杆菌Val98Ala RNHI 预计主要填充封闭状态。此处介绍的工作验证了该模型并确认了设计突变体的预测特性。MD 模拟表明手柄区域的构象偏好与 Trp85、Thr92 和 Val101 的构象相关。NMR 残余偶极耦合常数、三键标量耦合常数和化学位移通过实验定义了这些残基的构象状态,从而定义了手柄域的构象状态。这些 NMR 参数与 RNHI 同系物的 Michaelis 常数相关,证实了手柄区域在调节底物识别中的重要作用,并说明了 NMR 光谱在剖析酶功能的构象偏好方面的能力。



























 京公网安备 11010802027423号
京公网安备 11010802027423号