Journal of Allergy and Clinical Immunology ( IF 14.2 ) Pub Date : 2020-08-20 , DOI: 10.1016/j.jaci.2020.07.006 Anh N Do 1 , Yoojin Chun 1 , Galina Grishina 2 , Alexander Grishin 2 , Angela J Rogers 3 , Benjamin A Raby 4 , Scott T Weiss 5 , Alfin Vicencio 6 , Eric E Schadt 1 , Supinda Bunyavanich 7
|
Background
Nasal transcriptomics can provide an accessible window into asthma pathobiology.
Objective
Our goal was to move beyond gene signatures of asthma to identify master regulator genes that causally regulate genes associated with asthma phenotypes.
Methods
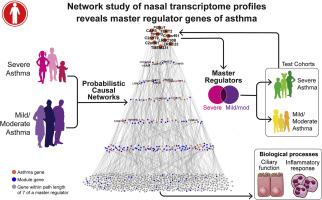
We recruited 156 children with severe persistent asthma and controls for nasal transcriptome profiling and applied network-based and probabilistic causal methods to identify severe asthma genes and their master regulators. We then took the same approach in an independent cohort of 190 adults with mild/moderate asthma and controls to identify mild/moderate asthma genes and their master regulators. Comparative analysis of the master regulator genes followed by validation testing in independent children with severe asthma (n = 21) and mild/moderate asthma (n = 154) was then performed.
Results
Nasal gene signatures for severe persistent asthma and for mild/moderate persistent asthma were identified; both were found to be enriched in coexpression network modules for ciliary function and inflammatory response. By applying probabilistic causal methods to these gene signatures and validation testing in independent cohorts, we identified (1) a master regulator gene common to asthma across severity and ages (FOXJ1); (2) master regulator genes of severe persistent asthma in children (LRRC23, TMEM231, CAPS, PTPRC, and FYB); and (3) master regulator genes of mild/moderate persistent asthma in children and adults (C1orf38 and FMNL1). The identified master regulators were statistically inferred to causally regulate the expression of downstream genes that modulate ciliary function and inflammatory response to influence asthma.
Conclusion
The identified master regulator genes of asthma provide a novel path forward to further uncovering asthma mechanisms and therapy.
中文翻译:

鼻转录组谱的网络研究揭示了哮喘的主要调节基因
背景
鼻转录组学可以为哮喘病理生物学提供一个可访问的窗口。
客观的
我们的目标是超越哮喘的基因特征,确定主要调节基因,这些基因可以因果调节与哮喘表型相关的基因。
方法
我们招募了 156 名患有严重持续性哮喘的儿童和对照组进行鼻转录组分析,并应用基于网络的概率因果方法来识别严重哮喘基因及其主要调节因子。然后,我们在一个由 190 名患有轻度/中度哮喘和对照的成年人组成的独立队列中采用了相同的方法,以确定轻度/中度哮喘基因及其主要调节因子。然后在患有严重哮喘(n = 21)和轻度/中度哮喘(n = 154)的独立儿童中对主要调节基因进行比较分析,然后进行验证测试。
结果
确定了严重持续性哮喘和轻度/中度持续性哮喘的鼻基因特征;两者都被发现富含纤毛功能和炎症反应的共表达网络模块。通过对这些基因特征和独立队列中的验证测试应用概率因果方法,我们确定了(1)不同严重程度和年龄的哮喘常见的主调节基因(FOXJ1);(2)儿童重症持续性哮喘主调控基因(LRRC23、TMEM231、CAPS、PTPRC、FYB);(3) 儿童和成人轻度/中度持续性哮喘的主调控基因(C1orf38和FMNL1). 统计推断确定的主要调节因子对调节纤毛功能和炎症反应以影响哮喘的下游基因的表达进行因果调节。
结论
已确定的哮喘主调节基因为进一步揭示哮喘机制和治疗提供了一条新途径。


























 京公网安备 11010802027423号
京公网安备 11010802027423号