Medicinal Chemistry ( IF 2.3 ) Pub Date : 2020-07-31 , DOI: 10.2174/1573406415666191004100744 Yina Wang 1 , Benrong Zheng 1 , Manbin Xu 2 , Shaoping Cai 3 , Jeong Younseo 4 , Chi Zhang 5 , Boxiong Jiang 1
|
Background: Renal cell carcinoma (RCC) is the most common malignant tumor of the adult kidney.
Objective: The aim of this study was to identify key genes signatures during RCC and uncover their potential mechanisms.
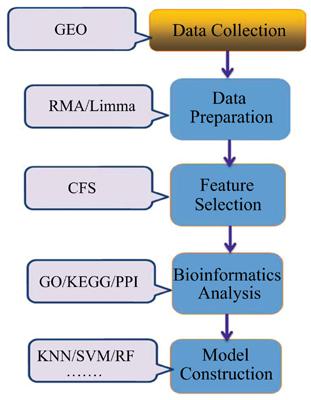
Methods: Firstly, the gene expression profiles of GSE53757 which contained 144 samples, including 72 kidney cancer samples and 72 controls, were downloaded from the GEO database. And then differentially expressed genes (DEGs) between the kidney cancer samples and the controls were identified. After that, GO and KEGG enrichment analyses of DEGs were performed by DAVID. Furthermore, the correlation-based feature subset (CFS) method was applied to the selection of key genes of DEGs. In addition, the classification model between the kidney cancer samples and the controls was built by Adaboost based on the selected key genes.
Results: 213 DEGs including 80 up-regulated and 133 down-regulated genes were selected as the feature genes to build the classification model between the kidney cancer samples and the controls by CFS method. The accuracy of the classification model by using 5-folds cross-validation test and independent set test is 84.4% and 83.3%, respectively. Besides, TYROBP, CD4163, CAV1, CXCL9, CXCL11 and CXCL13 also can be found in the top 20 hub genes screened by proteinprotein interaction (PPI) network.
Conclusion: It indicated that CFS is a useful tool to identify key genes in kidney cancer. Besides, we also predicted genes such as TYROBP, CD4163, CAV1, CXCL9, CXCL11 and CXCL13 that might target genes to diagnose the kidney cancer.
中文翻译:

基于CFS基因选择方法和Adaboost算法的肾细胞癌Hub基因预测与分析
背景:肾细胞癌(RCC)是成年肾脏最常见的恶性肿瘤。
目的:本研究的目的是鉴定RCC期间的关键基因特征并揭示其潜在机制。
方法:首先,从GEO数据库中下载了144个样本的GSE53757的基因表达谱,其中包括72个肾癌样本和72个对照。然后鉴定肾癌样品和对照之间的差异表达基因(DEG)。之后,DAVID对DEG进行了GO和KEGG富集分析。此外,基于相关的特征子集(CFS)方法被应用于DEGs关键基因的选择。此外,Adaboost基于选定的关键基因建立了肾癌样品与对照之间的分类模型。
结果:选择213个DEGs作为特征基因,包括80个上调基因和133个下调基因,以CFS方法建立肾癌标本与对照组的分类模型。使用5倍交叉验证检验和独立集检验的分类模型的准确性分别为84.4%和83.3%。此外,通过蛋白质-蛋白质相互作用(PPI)网络筛选的前20个集线器基因中也可以找到TYROBP,CD4163,CAV1,CXCL9,CXCL11和CXCL13。
结论:这表明CFS是鉴定肾癌关键基因的有用工具。此外,我们还预测了TYROBP,CD4163,CAV1,CXCL9,CXCL11和CXCL13等基因可能成为诊断肾脏癌的靶基因。


























 京公网安备 11010802027423号
京公网安备 11010802027423号