Journal of Pharmaceutical Analysis ( IF 8.8 ) Pub Date : 2020-08-02 , DOI: 10.1016/j.jpha.2020.07.008 Xinyue Yu 1, 2 , Jingxue Nai 1 , Huimin Guo 3 , Xuping Yang 1 , Xiaoying Deng 1 , Xia Yuan 1 , Yunfei Hua 1 , Yuan Tian 1 , Fengguo Xu 1 , Zunjian Zhang 1 , Yin Huang 1, 2
|
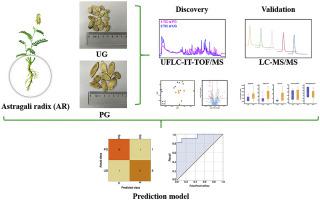
Astragali radix (AR, the dried root of Astragalus) is a popular herbal remedy in both China and the United States. The commercially available AR is commonly classified into premium graded (PG) and ungraded (UG) ones only according to the appearance. To uncover novel sensitive and specific markers for AR grading, we took the integrated mass spectrometry-based untargeted and targeted metabolomics approaches to characterize chemical features of PG and UG samples in a discovery set (n=16 batches). A series of five differential compounds were screened out by univariate statistical analysis, including arginine, calycosin, ononin, formononetin, and astragaloside Ⅳ, most of which were observed to be accumulated in PG samples except for astragaloside Ⅳ. Then, we performed machine learning on the quantification data of five compounds and constructed a logistic regression prediction model. Finally, the external validation in an independent validation set of AR (n=20 batches) verified that the five compounds, as well as the model, had strong capability to distinguish the two grades of AR, with the prediction accuracy > 90%. Our findings present a panel of meaningful candidate markers that would significantly catalyze the innovation in AR grading.



























 京公网安备 11010802027423号
京公网安备 11010802027423号