Cellular and Molecular Gastroenterology and Hepatology ( IF 7.2 ) Pub Date : 2020-07-31 , DOI: 10.1016/j.jcmgh.2020.07.010 Valeriia Dotsenko 1 , Mikko Oittinen 1 , Juha Taavela 2 , Alina Popp 3 , Markku Peräaho 4 , Synnöve Staff 5 , Jani Sarin 6 , Francisco Leon 7 , Jorma Isola 6 , Markku Mäki 1 , Keijo Viiri 1
|
Background & Aims
Gluten challenge studies are instrumental in understanding the pathophysiology of celiac disease. Our aims in this study were to reveal early gluten-induced transcriptomic changes in duodenal biopsies and to find tools for clinics.
Methods
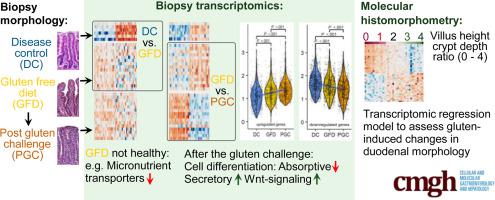
Duodenal biopsies were collected from 15 celiac disease patients on a strict long-term gluten-free diet (GFD) prior to and post a gluten challenge (PGC) and from 6 healthy control individuals (DC). Biopsy RNA was subjected to genome-wide 3’ RNA-Seq. Sequencing data was used to determine the differences between the three groups and was compared to sequencing data from the public repositories. The biopsies underwent morphometric analyses.
Results
In DC vs. GFD group comparisons, 167 differentially expressed genes were identified with 117 genes downregulated and 50 genes upregulated. In PGC vs. GFD group comparisons, 417 differentially expressed genes were identified with 195 genes downregulated and 222 genes upregulated. Celiac disease patients on a GFD were not “healthy”. In particular, genes encoding proteins for transporting small molecules were expressed less. In addition to the activation of immune response genes, a gluten challenge induced hyperactive intestinal wnt-signaling and consequent immature crypt gene expression resulting in less differentiated epithelium. Biopsy gene expression in response to a gluten challenge correlated with the extent of the histological damage. Regression models using only four gene transcripts described 97.2% of the mucosal morphology and 98.0% of the inflammatory changes observed.
Conclusions
Our gluten challenge trial design provided an opportunity to study the transition from health to disease. The results show that even on a strict GFD, despite being deemed healthy, patients reveal patterns of ongoing disease. Here, a transcriptomic regression model estimating the extent of gluten-induced duodenal mucosal injury is presented.
中文翻译:

无麸质饮食和后麸质挑战的乳糜泻患者肠道粘膜的全基因组转录组学分析。
背景与目标
麸质挑战研究有助于了解乳糜泻的病理生理学。我们在这项研究中的目的是揭示十二指肠活检中早期麸质诱导的转录组变化,并为临床寻找工具。
方法
从 15 名接受麸质挑战 (PGC) 之前和之后严格长期无麸质饮食 (GFD) 的乳糜泻患者和 6 名健康对照个体 (DC) 收集十二指肠活检组织。活检 RNA 进行全基因组 3' RNA-Seq。测序数据用于确定三组之间的差异,并与来自公共存储库的测序数据进行比较。活检进行了形态测量分析。
结果
在 DC 与 GFD 组比较中,鉴定出 167 个差异表达基因,其中 117 个基因下调,50 个基因上调。在 PGC 与 GFD 组比较中,鉴定出 417 个差异表达基因,其中 195 个基因下调,222 个基因上调。GFD 上的乳糜泻患者并不“健康”。特别是,编码用于运输小分子的蛋白质的基因表达较少。除了激活免疫反应基因外,麸质挑战还会诱导过度活跃的肠道 wnt 信号传导和随之而来的不成熟的隐窝基因表达,导致上皮分化程度降低。响应面筋挑战的活检基因表达与组织学损伤的程度相关。仅使用四个基因转录本的回归模型描述了 97.2% 的粘膜形态和 98%。
结论
我们的麸质挑战试验设计为研究从健康到疾病的转变提供了机会。结果表明,即使在严格的 GFD 下,尽管患者被认为是健康的,但仍会显示出持续疾病的模式。在这里,介绍了估计麸质诱导的十二指肠粘膜损伤程度的转录组学回归模型。


























 京公网安备 11010802027423号
京公网安备 11010802027423号