Cell ( IF 64.5 ) Pub Date : 2020-07-30 , DOI: 10.1016/j.cell.2020.07.003 Sydney M Shaffer 1 , Benjamin L Emert 2 , Raúl A Reyes Hueros 3 , Christopher Cote 4 , Guillaume Harmange 5 , Dylan L Schaff 6 , Ann E Sizemore 6 , Rohit Gupte 6 , Eduardo Torre 3 , Abhyudai Singh 7 , Danielle S Bassett 8 , Arjun Raj 4
|
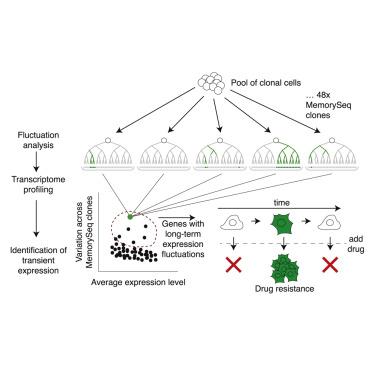
Non-genetic factors can cause individual cells to fluctuate substantially in gene expression levels over time. It remains unclear whether these fluctuations can persist for much longer than the time of one cell division. Current methods for measuring gene expression in single cells mostly rely on single time point measurements, making the duration of gene expression fluctuations or cellular memory difficult to measure. Here, we combined Luria and Delbrück’s fluctuation analysis with population-based RNA sequencing (MemorySeq) for identifying genes transcriptome-wide whose fluctuations persist for several divisions. MemorySeq revealed multiple gene modules that expressed together in rare cells within otherwise homogeneous clonal populations. These rare cell subpopulations were associated with biologically distinct behaviors like proliferation in the face of anti-cancer therapeutics. The identification of non-genetic, multigenerational fluctuations can reveal new forms of biological memory in single cells and suggests that non-genetic heritability of cellular state may be a quantitative property.


























 京公网安备 11010802027423号
京公网安备 11010802027423号