Journal of Theoretical Biology ( IF 2 ) Pub Date : 2020-07-23 , DOI: 10.1016/j.jtbi.2020.110414 Wei Liu 1 , Liangyu Ma 2 , Ling Chen 2 , Bolun Chen 3 , Byeungwoo Jeon 4 , Jipeng Qiang 2
|
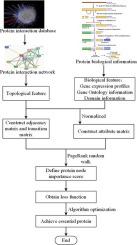
Mining essential protein is crucial for discovering the process of cellular organization and viability. At present, there are many computational methods for essential proteins detecting. However, these existing methods only focus on the topological information of the networks and ignore the biological information of proteins, which lead to low accuracy of essential protein identification. Therefore, this paper presents a new essential proteins prediction strategy, called DEP-MSB which integrates a variety of biological information including gene expression profiles, GO annotations, and Domain interaction strength. In order to evaluate the performance of DEP-MSB, we conduct a series of experiments on the yeast PPI network and the experimental results have shown that the proposed algorithm DEP-MSB is more superior to the other existing traditional methods and has obviously improvement in prediction accuracy.
中文翻译:

一种基于多源生物信息的必需蛋白发现新方案。
挖掘必需蛋白质对于发现细胞组织和生存能力至关重要。目前,有许多用于检测必需蛋白质的计算方法。但是,这些现有方法仅关注网络的拓扑信息,而忽略了蛋白质的生物学信息,这导致必需蛋白质鉴定的准确性较低。因此,本文提出了一种新的必需蛋白质预测策略,称为DEP-MSB,该策略整合了包括基因表达谱,GO注释和域相互作用强度在内的多种生物学信息。为了评估DEP-MSB的性能,


























 京公网安备 11010802027423号
京公网安备 11010802027423号