Journal of Molecular Graphics and Modelling ( IF 2.9 ) Pub Date : 2020-07-15 , DOI: 10.1016/j.jmgm.2020.107695 Koichiro Kato 1 , Teruki Honma 2 , Kaori Fukuzawa 3
|
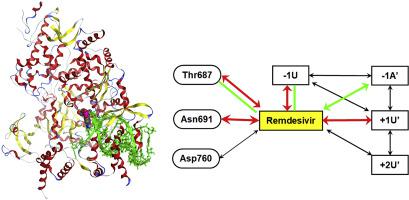
COVID-19, a disease caused by a new strain of coronavirus (SARS-CoV-2) originating from Wuhan, China, has now spread around the world, triggering a global pandemic, leaving the public eagerly awaiting the development of a specific medicine and vaccine. In response, aggressive efforts are underway around the world to overcome COVID-19. In this study, referencing the data published on the Protein Data Bank (PDB ID: 7BV2) on April 22, we conducted a detailed analysis of the interaction between the complex structures of the RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 and Remdesivir, an antiviral drug, from the quantum chemical perspective based on the fragment molecular orbital (FMO) method. In addition to the hydrogen bonding and intra-strand stacking between complementary strands as seen in normal base pairs, Remdesivir bound to the terminus of an primer-RNA strand was further stabilized by diagonal π-π stacking with the -1A’ base of the complementary strand and an additional hydrogen bond with an intra-strand base, due to the effect of chemically modified functional group. Moreover, stable OH/π interaction is also formed with Thr687 of the RdRp. We quantitatively revealed the exhaustive interaction within the complex among Remdesivir, template-primer-RNA, RdRp and co-factors, and published the results in the FMODB database.


























 京公网安备 11010802027423号
京公网安备 11010802027423号