Journal of Molecular Graphics and Modelling ( IF 2.9 ) Pub Date : 2020-07-09 , DOI: 10.1016/j.jmgm.2020.107650 Shigenori Tanaka 1 , Chiduru Watanabe 2 , Teruki Honma 3 , Kaori Fukuzawa 4 , Kazue Ohishi 5 , Tadashi Maruyama 6
|
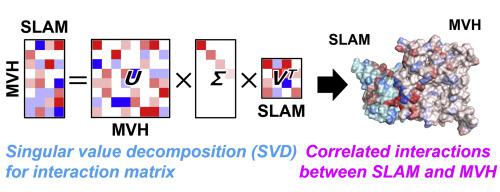
A theoretical scheme to systematically describe correlated (network-like) interactions between molecular fragments is proposed within the framework of the fragment molecular orbital (FMO) method. The method is mathematically based on the singular value decomposition (SVD) of the inter-fragment interaction energy (IFIE) matrix obtained by the FMO calculation, and can be applied to a comprehensive description of protein-protein interactions in the context of molecular recognition. In the present study we apply the proposed method to a complex of measles virus hemagglutinin and human SLAM receptor, thus finding a usefulness for efficiently eliciting the correlated interactions among the amino-acid residues involved in the two proteins. Additionally, collective interaction networks by amino-acid residues important for mutation experiments can be clearly visualized.
中文翻译:

基于片段分子轨道法鉴定蛋白质复合物中相关残基间相互作用。
在片段分子轨道(FMO)方法的框架内提出了一种系统地描述分子片段之间相关(网络状)相互作用的理论方案。该方法在数学上基于FMO计算得到的片段间相互作用能(IFIE)矩阵的奇异值分解(SVD),可应用于分子识别背景下蛋白质-蛋白质相互作用的综合描述。在本研究中,我们将所提出的方法应用于麻疹病毒血凝素和人类 SLAM 受体的复合物,从而发现了有效引发两种蛋白质所涉及的氨基酸残基之间相关相互作用的有用性。此外,对于突变实验很重要的氨基酸残基的集体相互作用网络可以清晰地可视化。



























 京公网安备 11010802027423号
京公网安备 11010802027423号