Cell Chemical Biology ( IF 8.6 ) Pub Date : 2020-07-09 , DOI: 10.1016/j.chembiol.2020.06.014 Zachary E Potter 1 , Ho-Tak Lau 2 , Sujata Chakraborty 1 , Linglan Fang 1 , Miklos Guttman 3 , Shao-En Ong 2 , Douglas M Fowler 4 , Dustin J Maly 5
|
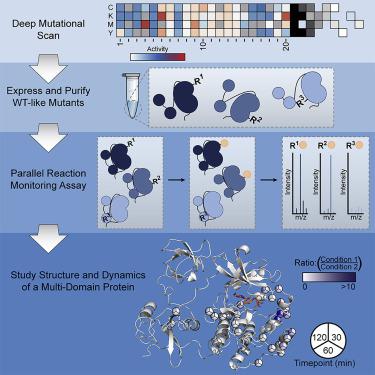
Solution-based structural techniques complement high-resolution structural data by providing insight into the oft-missed links between protein structure and dynamics. Here, we present Parallel Chemoselective Profiling, a solution-based structural method for characterizing protein structure and dynamics. Our method utilizes deep mutational scanning saturation mutagenesis data to install amino acid residues with specific chemistries at defined positions on the solvent-exposed surface of a protein. Differences in the extent of labeling of installed mutant residues are quantified using targeted mass spectrometry, reporting on each residue's local environment and structural dynamics. Using our method, we studied how conformation-selective, ATP-competitive inhibitors affect the local and global structure and dynamics of full-length Src kinase. Our results highlight how parallel chemoselective profiling can be used to study a dynamic multi-domain protein, and suggest that our method will be a useful addition to the relatively small toolkit of existing protein footprinting techniques.
中文翻译:

用于绘制蛋白质结构的并行化学选择性分析。
基于解决方案的结构技术通过提供对蛋白质结构和动力学之间经常被忽略的联系的洞察来补充高分辨率结构数据。在这里,我们介绍了 Parallel Chemoselective Profiling,这是一种基于溶液的结构方法,用于表征蛋白质结构和动力学。我们的方法利用深度突变扫描饱和诱变数据在蛋白质的溶剂暴露表面上的定义位置安装具有特定化学性质的氨基酸残基。使用靶向质谱法对安装的突变残留物标记程度的差异进行量化,报告每个残留物的局部环境和结构动力学。使用我们的方法,我们研究了构象选择性、ATP 竞争性抑制剂如何影响全长 Src 激酶的局部和全局结构和动力学。



























 京公网安备 11010802027423号
京公网安备 11010802027423号